We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

In living organisms, all cells carry the same genomic DNA, yet each cell fulfils a unique function by selectively activating or repressing the expression of specific genes. This process relies on the precise regulation of thousands of regulatory proteins that localize to specific regions of DNA and perform the task of activating or repressing transcription.
The field of molecular biology has historically been concerned with elucidating the interactions between proteins and DNA sequences, and the manner in which they collaborate to regulate cell-specific gene expression. In order to study these complex regulatory mechanisms, scientists have developed advanced techniques such as ChIP-seq and CUT&Tag. However, these techniques typically analyse individual proteins and primarily focus on histone modifications and high abundance proteins. This is due to the fact that, given the sheer multitude of regulatory proteins and their interactions within the gene regulatory network, it remains a challenge to construct a comprehensive regulatory factor map in order to achieve a more in-depth analysis of the intricacies of gene regulation.
A team of researchers from the California Institute of Technology (Caltech) and other institutions recently collaborated to publish an article entitled "ChIP-DIP maps binding of hundreds of proteins to DNA simultaneously and identifies ChIP-DIP maps binding of hundreds of proteins to DNA simultaneously and identifies diverse gene regulatory elements" in Nature Genetics. The article reports the development of a newly developed parallel chromatin immunoprecipitation assay, ChIP-DIP.
The technology has the capacity to detect the binding of hundreds of regulatory proteins to DNA in a single experiment. This process generates a genomic atlas comprising hundreds of different regulatory proteins, thereby providing a more comprehensive view of the dynamics of gene regulation. ChIP-DIP has been shown to generate highly accurate maps for a wide range of DNA-associated proteins, including modified histones, chromatin regulators, and transcription factors. The technique has been demonstrated to be suitable for use in different experimental conditions and molecular biology platforms, which can help to generate context-specific protein localization maps on a federated scale in multiple laboratories and experimental systems. This has the potential to reveal the complex details of gene regulation.
Take the Next Step: Explore Related Services
The research team developed a technique known as ChIP-DIP, which enables the simultaneous revelation of the various effects of multiple proteins on DNA in a single experiment. This technique involves the combination of multiple antibodies with magnetic beads, along with unique oligonucleotide tags, to form a pool of antibody-beads. The subsequent performance of ChIP assigns a unique identifier to each antibody-DNA complex, facilitating their analysis in subsequent studies. The method generates localization profiles for each protein by means of DNA read clustering with the same barcode paired with the same antibody through high-throughput sequencing.
In order to evaluate and compare the ChIP-DIP localization accuracy, the research team used four typical proteins in human K562 cells. The results demonstrate that, in terms of resolving protein localization, ChIP-DIP is essentially consistent with the traditional classical ChIP-seq method and exhibits good accuracy, efficiency and robustness. Notably, ChIP-DIP overcomes the limitations of conventional ChIP technology by facilitating the simultaneous detection of protein-DNA interactions, including those involving low-abundance proteins.
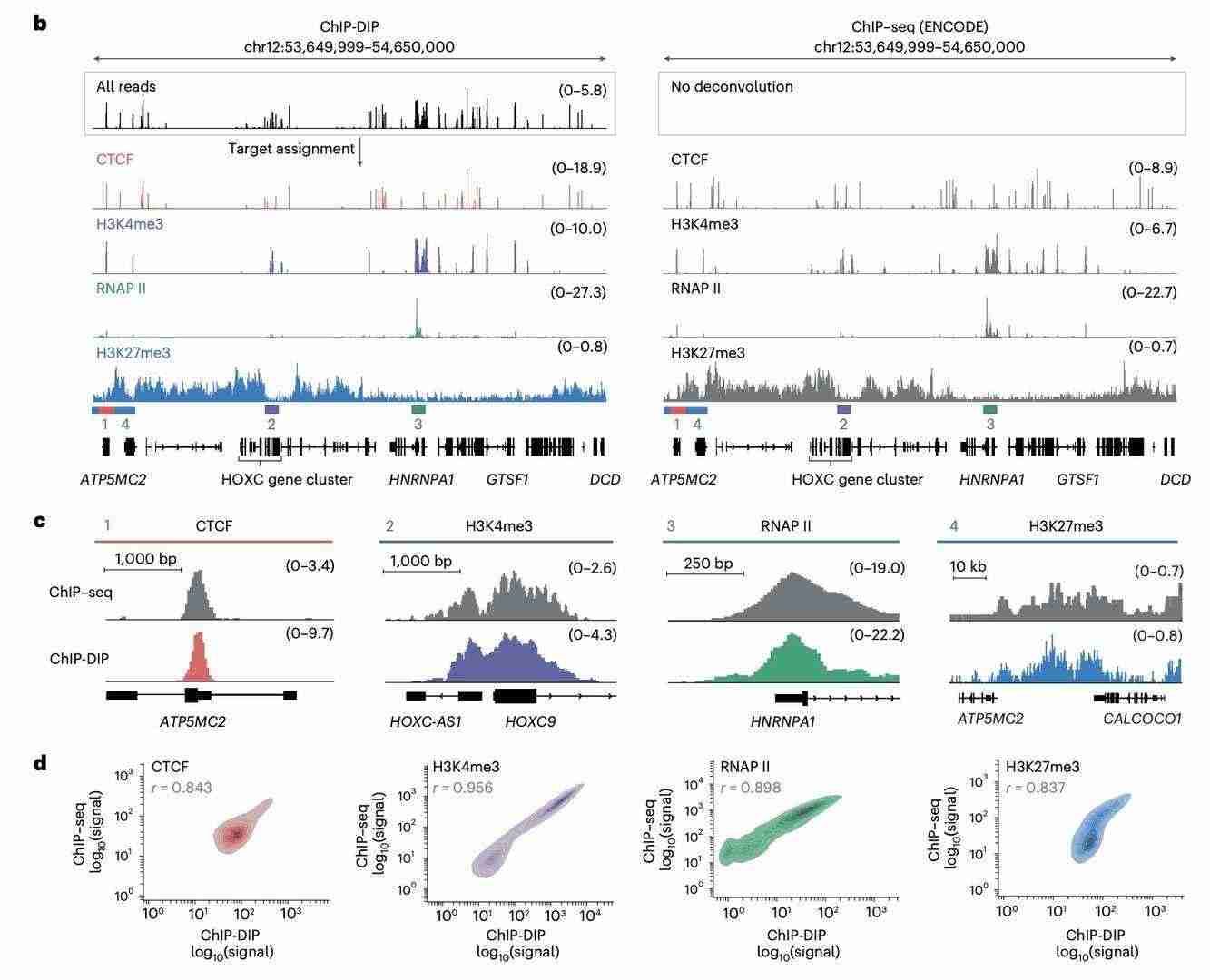 ChIP-DIP accurately localizes proteins to genomic DNA regions (Perez et al., 2024)
ChIP-DIP accurately localizes proteins to genomic DNA regions (Perez et al., 2024)
ChIP-DIP is a powerful tool for efficiently identifying genomic regions that bind to specific antibodies and constructing accurate protein localization maps. It combines immunoprecipitation and molecular barcoding technologies.First, antibodies are coupled to magnetic beads and labeled by oligonucleotide tags to ensure stable binding between the antibody and the beads and subsequent tracking by the tags. This stage sets the stage for subsequent experiments. Subsequently, multiple antibody-magnetic bead-oligo complexes are amalgamated to constitute a pool of antibody-magnetic beads, which are capable of specifically binding to target DNA fragments on chromosomes, thereby completing the chromatin immunoprecipitation (ChIP) process.
Subsequent to the completion of the ChIP process, a series of repeated splitting and pooling barcoding operations are conducted. This step guarantees precise categorisation and tracking of DNA fragments by assigning a unique barcode to each chromatin-antibody-magnetic bead-oligonucleotide complex. This process enhances the precision of the operation and furnishes the requisite information for subsequent high-throughput sequencing analysis. Subsequently, the DNA fragments are subjected to high-throughput sequencing, and the barcode information of the genomic DNA and antibody-oligonucleotide from the sequencing results are clustered to assign a corresponding antibody to each genomic region.
Finally, by summarising all DNA sequence reads associated with a particular antibody, protein localisation maps can be generated. These localisation maps reveal the interaction patterns between different antibodies and genomic regions, providing valuable insights into chromatin modifications, transcription factor binding, and other genomic functions.The ChIP-DIP approach, by fusing chromatin immunoprecipitation with barcoding technology, allows us to reveal chromatin structure and genomic regulatory mechanisms at a higher resolution.
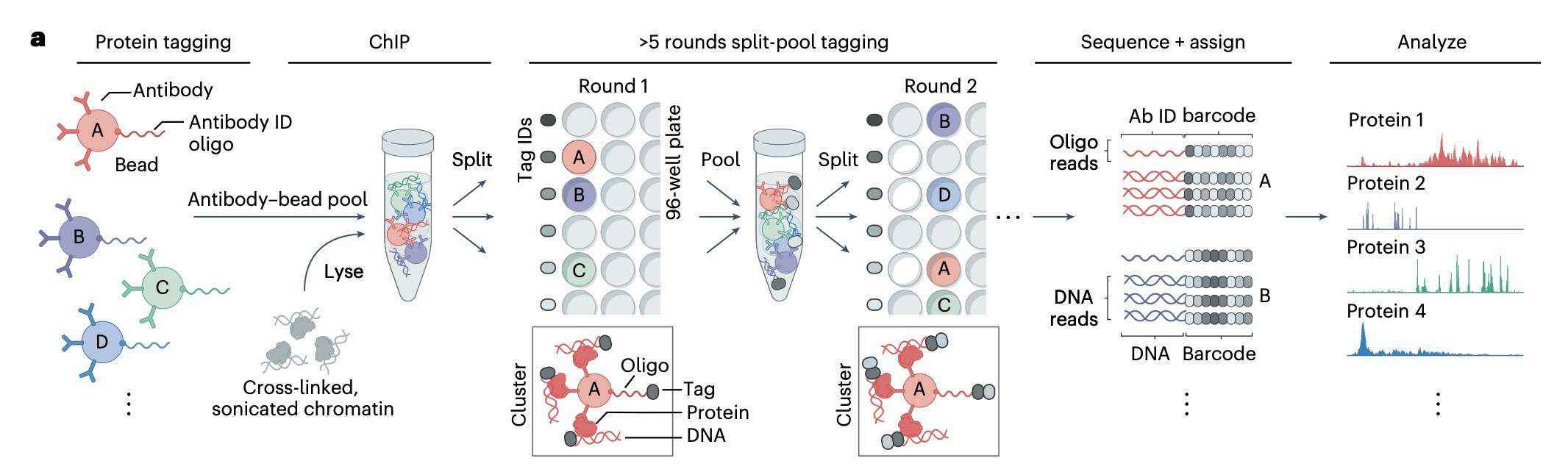 Schematic of the experimental design (Perez et al., 2024)
Schematic of the experimental design (Perez et al., 2024)
ChIP-DIP technology is not only suitable for static gene regulation studies, but also capable of revealing the dynamic process of gene regulation. Utilising mouse bone marrow dendritic cells (mDCs), the research team induced antimicrobial pathogen responses by lipopolysaccharide (LPS) treatment, and analysed the dynamic changes in protein localization under a variety of experimental conditions using ChIP-DIP. The results demonstrated that different chromatin modifications changed significantly at different time points, but the patterns of temporal changes varied. For the enriched H3K27ac modification region, the changes were in three patterns: stable state, activated state and repressed state, which corresponded to gene transcription remaining unchanged, increasing and decreasing, respectively.
Furthermore, while both promoter and enhancer chromatin modifications change in response to LPS stimulation, enhancer modifications are more consistent with changes in transcriptional activity.These results not only reveal that immune cells finely regulate gene expression through dynamic changes in chromatin, but also indicate that ChIP-DIP is capable of directly characterising changes in protein localization in different samples, time points, or a variety of biological samples in response to perturbation.
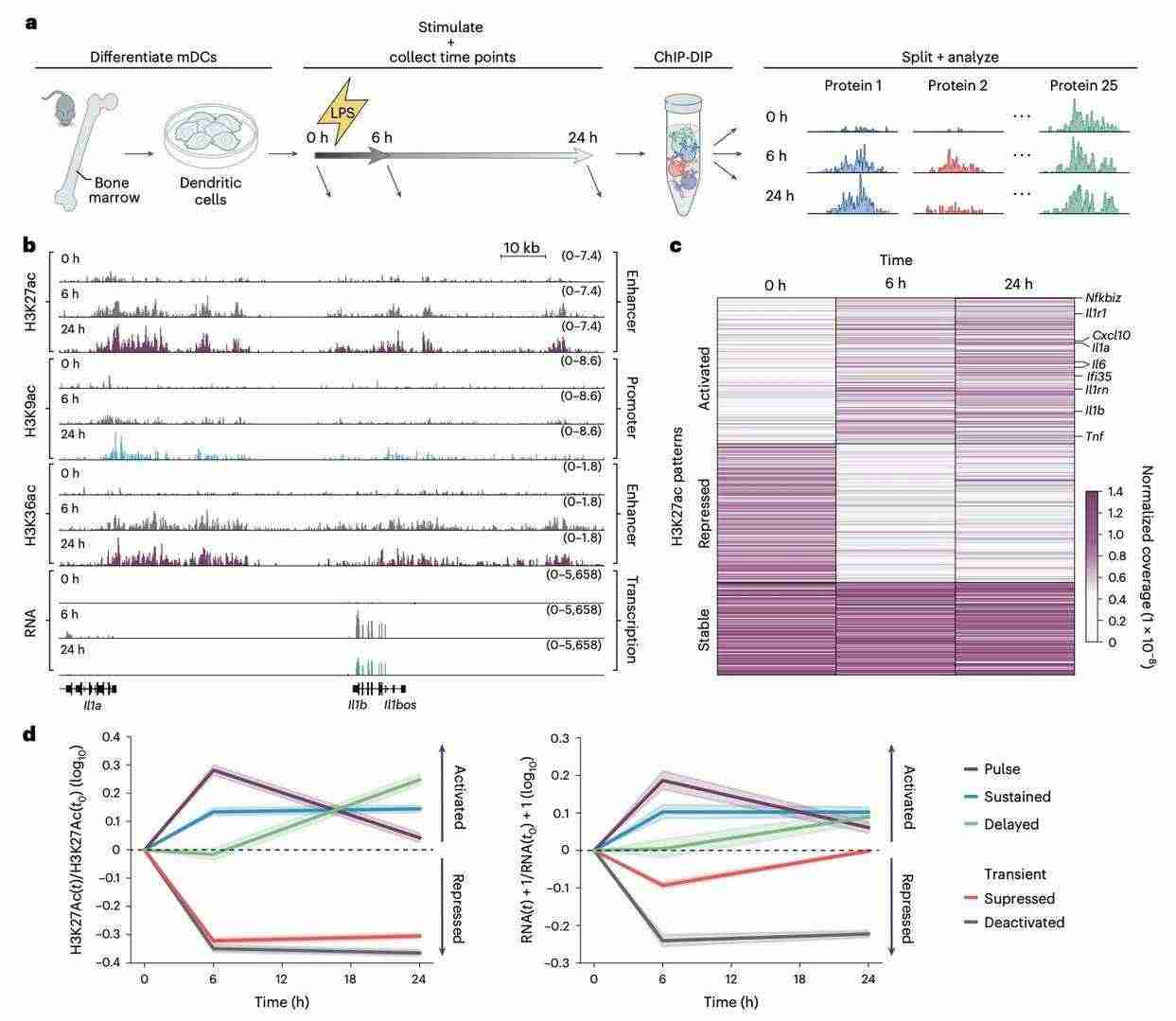 ChIP-DIP reveals dynamic changes in chromatin profiles after LPS stimulation of mDCs (Perez et al., 2024)
ChIP-DIP reveals dynamic changes in chromatin profiles after LPS stimulation of mDCs (Perez et al., 2024)
Previous studies have identified the role of histone modifications in distinguishing genomic elements and their active states, but most have focused on five common histone modifications. To expand the understanding of other histone modifications and regulatory proteins, the research team performed in-depth analysis using ChIP-DIP technology.
It was determined that H3K4me3 is predominantly present on the promoters of actively transcribed RNAP II genes, yet it is also observed in the vicinity of RNAP I promoters and active RNAP III genes. Furthermore, other histone modifications associated with active RNAP II promoters are enriched on RNAP I and III genes. Additionally, the level of histone modifications and transcriptional start sites varies by RNAP gene. Given this property, promoter characteristics can be distinguished by different combinations of histone modifications, including polymerase, gene type, and activity level.
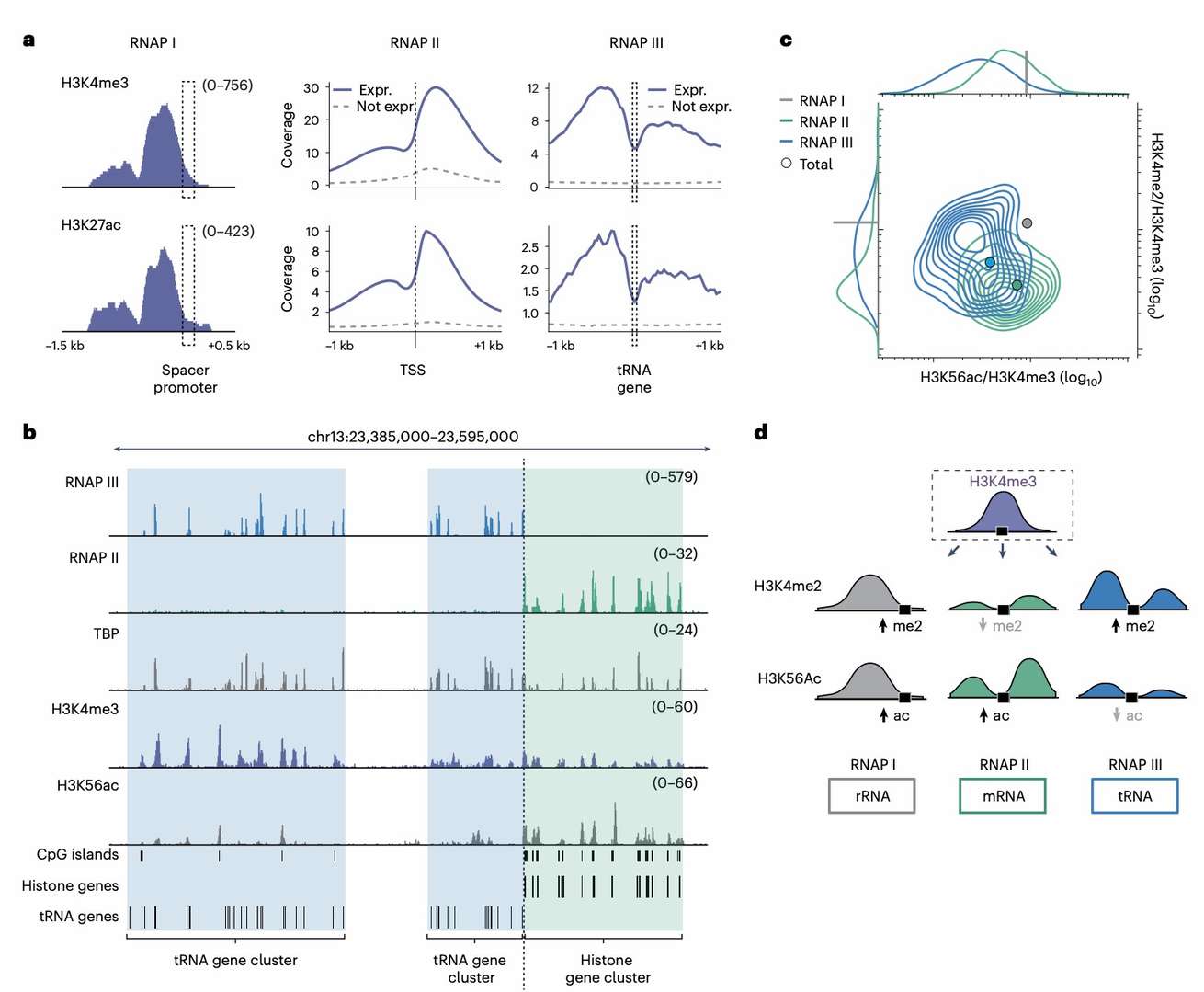 Distinct chromatin signatures define the promoters of each RNAP (Perez et al., 2024)
Distinct chromatin signatures define the promoters of each RNAP (Perez et al., 2024)
ChIP-DIP has been demonstrated to exhibit both a high level of proficiency in small-scale experiments and an equally impressive capacity for scalability. Through the manipulation of antibody pools in terms of size and combination, researchers are able to analyse a substantial number of protein-DNA interactions within a single experiment.A notable illustration of this is the K562 cell experiment, in which 52 antibodies were employed, resulting in the successful capture of over 160 distinct proteins, encompassing transcription factors, histone modifications, and chromatin regulators. This capacity renders ChIP-DIP a potent instrument for the exploration of gene regulatory networks.
Furthermore, the high sensitivity of the ChIP-DIP technology ensures the generation of reliable data from smaller cell samples.In their experiments, the researchers utilised cell volumes ranging from 50,000 cells to 4.5 million cells, and the results demonstrated the capacity of ChIP-DIP to successfully generate localization profiles for 35 distinct proteins with a mere 50,000 cells. This finding indicates that ChIP-DIP is not only suitable for common large-scale cell samples, but can also be applied to analyse protein-DNA interactions in rare cell types or clinical samples.
This study introduces ChIP-DIP, a pioneering technological innovation that facilitates the simultaneous detection of numerous protein-DNA interactions, thereby markedly enhancing the efficiency of research endeavours.
ChIP-DIP technology represents a significant advancement in the field of molecular biology by overcoming the limitations of conventional ChIP technology. This technology employs parallel immunoprecipitation and high-throughput sequencing, enabling the analysis of multiple protein-DNA interactions in a single experiment. The technology's core strengths are rooted in the establishment of multiplexed antibody pools, the implementation of split-pool barcoding technology, and the precise localization facilitated by high-throughput sequencing. These innovative steps ensure independent labelling and precise differentiation of different protein-DNA binding events, thus improving the efficiency, sensitivity and accuracy of the experiment.
ChIP-DIP technology has been demonstrated to be capable of analysing different types of proteins, including transcription factors and histone modifications, in a simultaneous manner. In addition to this, it provides a novel perspective on the distribution and interaction of proteins within the genome, thereby promoting in-depth exploration of gene regulation mechanisms. Conventional molecular biology techniques are employed, facilitating its application in diverse research settings. The technique is straightforward to operate and does not necessitate specialised equipment or training, making it a promising key tool for gene regulation analysis. With ongoing technological refinement and broadening of its applications, ChIP-DIP is poised to assume an instrumental role in disease research.
Reference

CD Genomics is transforming biomedical potential into precision insights through seamless sequencing and advanced bioinformatics.
We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy