CD Genomics offers a rapid and accurate custom SNP validation on the MassARRAY MALDI–TOF instrument provided by Agena Bioscience. Our MassARRAY SNP Genotyping system combines mass spectrometry, sensitive and robust chemistry, and advanced data analysis software to meet your SNP genotyping needs.
Introduction of MassARRAY SNP Genotyping
MassARRAY SNP Genotyping amplifies the DNA sequence across the SNP site by PCR, and then uses a single extension primer to amplify PCR products, ensuring that the primer extends only one base. The extended product was analyzed by TOF (time of flight) and the SNP was classified according to the molecular weight difference of base, and the allele frequency of the SNP site could also be calculated. MassARRAY SNP genotyping can carry out multiple SNP typing in one reaction. This technique is accurate and has a price advantage when analyzing dozens or hundreds of SNPs.
With different detection and software modules, the MassARRAY® iPLEX GOLD technology can realize the research or application of SNP genotyping, gene expression research, copy number variation, gene methylation analysis, pathogen typing and prenatal diagnosis on one platform. MassARRAY SNP Genotyping has a very good application prospect in the fields of biological science, medicine, agricultural research and so on. It has provided excellent solutions for scientists all over the world.
Advantages of MassARRAY SNP Genotyping
- Accurate and Automated: Nucleic acid detection by mass spectrometry enables readout by molecular mass, and using state-of-the-art technologies for automated, non-contact dispensing of reagents.
- Scalable and Flexible: We offer a variety of options for SNP, sample and assay throughput.
- High Multiplex Capability: Multiplex up to 30 SNP loci in a single pool.
- Fast Assay Design: A new assay can be designed within a few hours. In addition, a semi-automated workflow and unmodified oligos provide rapid turnaround time.
- Cost Effective: Multiplex analysis reduces per-sample cost, and scalable throughput optimizes batch and resource requirements.
Applications of MassARRAY SNP Genotyping
- Genotyping and mutation detection
- Ultrasensitive detection
- Methylation analysis
- Variety identification
- Functional Panel customization
- Pharmacogenomics research
- Biomarker validation
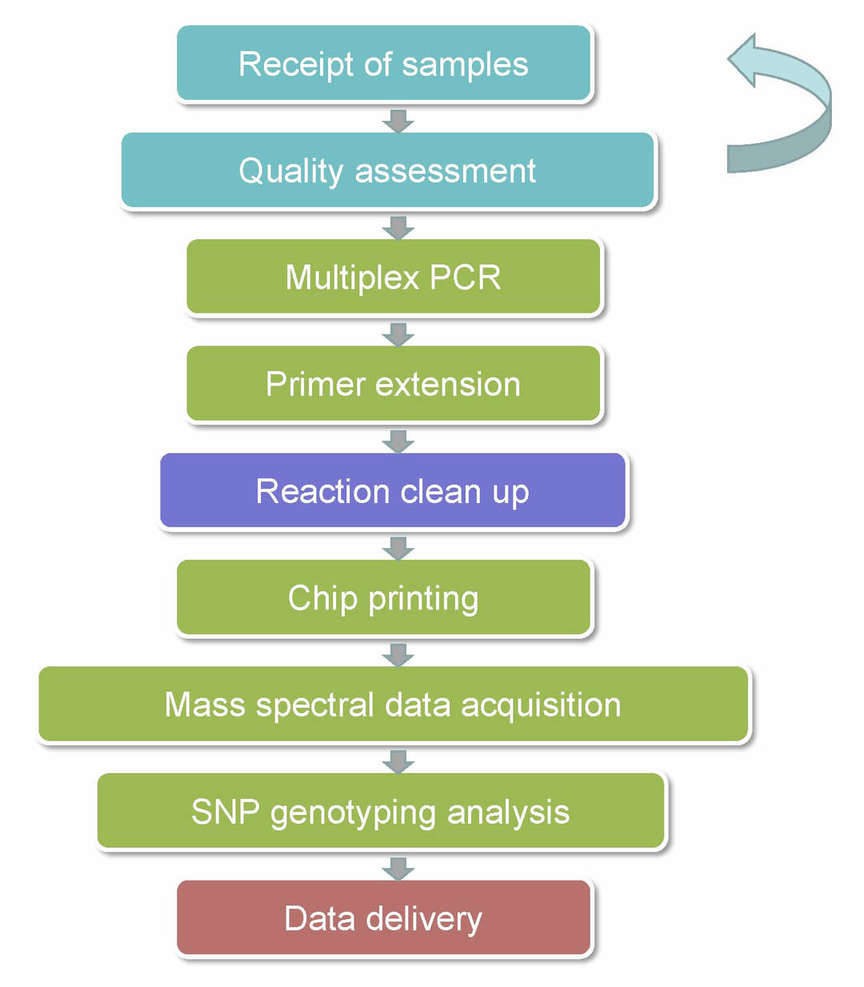
MassARRAY SNP Genotyping Workflow
The procedures for our MassARRAY SNP Genotyping is illustrated below.
Service Specifications
Sample requirements
|
|
| Chip Printing | |
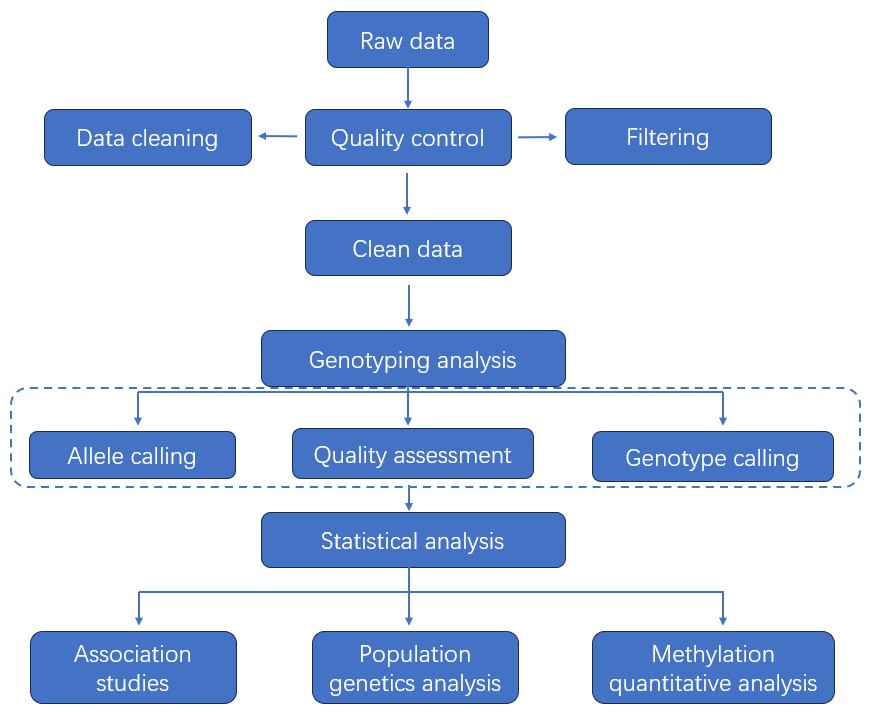
Data Analysis
|
Analysis Pipeline
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
Our service includes customized SNP panel design, oligo manufacture and pool balancing, iPLEX chemistry running, analysis of genotyping, and delivery of genotype report in Excel format. Our workflows can be designed to accommodate both small and large assay requirements. If you have additional requirements or questions, please feel free to contact us.
References:
- Aierken K, Dong Z, Abulimiti T, et al. CDK6 3'UTR polymorphisms alter the susceptibility to cervical cancer among Uyghur females. Mol Genet Genomic Med. 2019 Mar 4:e626.
- Cai J, Cui K, Niu F, et al. Genetics of IL6 polymorphisms: Case-control study of the risk of endometrial cancer. Mol Genet Genomic Med. 2019 Mar 3:e600.
- Ji H, Lu L, Huang J, et al. IL1A polymorphisms is a risk factor for colorectal cancer in Chinese Han population: a case control study. BMC Cancer. 2019 Feb 28;19(1):181.
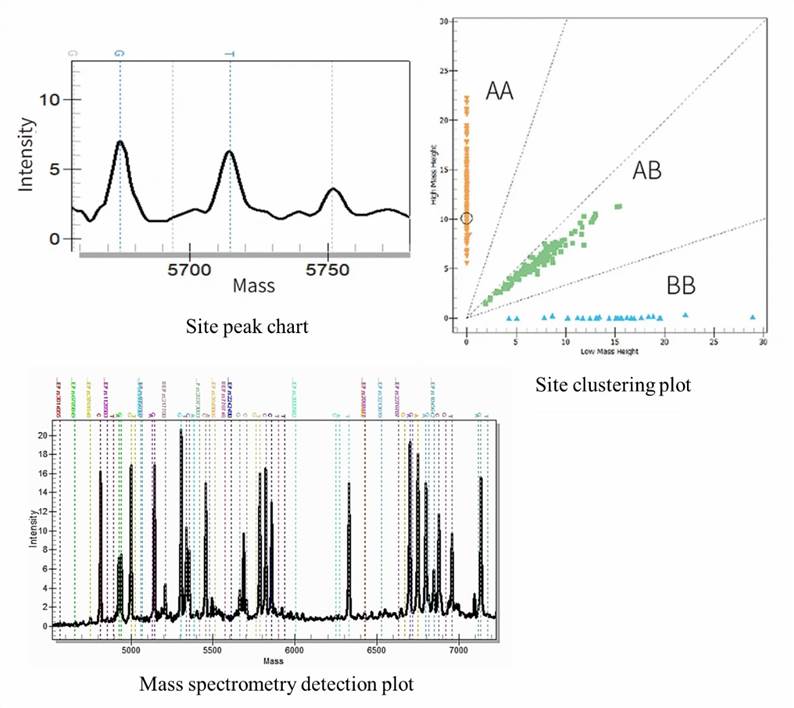
1. How many samples can be processed on the MassARRAY System?
It depends on the assay design and the format of the SpectroCHIP Array being utilized. The MassARRAY System processes 2 chips per run. The chip comes in one of two formats, 96- or 384-well format. If the assay design calls for a single well, the number of samples processed in a single run ranges from 768 samples with the 384-well format chips to 192 samples with the 96-well format.
2. How accurate and reproducible are the experimental results of Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF)?
Under certain experimental conditions, the accuracy and reproducibility of SNP genotyping detection results can both reach above 99%.
3. How reliable is the MassARRAY SNP Assay?
Developed by the American company Sequenom, the MassARRAY technology has found a wide array of applications in disease research, molecular diagnostics, and molecular breeding. Additionally, research utilizing this technology has been published in reputable scientific journals such as Nature, Science, and Blood.
4. What is the principle of MassARRAY?
The MassARRAY gene analysis technology is predicated upon the MALDT-TOF mass spectrometry technique. The analytical principle of this technology hinges on identifying base variations at specific SNP locations via the weight difference of individual bases. Further, it exhibits the capability to detect DNA within the approximate mass range of 4500da to 9000Da, with a mass separation resolution of 16Da.
Variation in the glucose transporter gene SLC2A2 is associated with glycemic response to metformin
Journal: Nature genetics
Impact factor: 27.125
Published: 08 August 2016
Backgrounds
Metformin, a quintessential antihyperglycemic agent, is administered to over 100 million users globally. Nevertheless, its mechanism of action is not yet fully elucidated. Apart from diabetes, metformin also provides therapeutic benefits to other pathological conditions such as fatty liver diseases and Parkinson's disease. However, a considerable segment of individuals diagnosed with Type 2 diabetes does not respond effectively to the usual dosage of metformin. The authors, in this regard, have undertaken a meticulous investigation through the screening of diverse samples. Utilizing techniques such as Genome-Wide Association Studies (GWAS) and MassArray SNP genotyping, they aim to identify specific genetic markers, thus providing a valuable guide for precision medicine.
Methods
- First stage: 1,473 samples
- Second stage: 1,223 samples
- Third stage: 10,557 samples of European ancestry
- and 2,566 samples of non-European ancestry
- GWAS screening
- MassArray SNP genotyping
- TaqMan-based Allelic Discrimination assays
- Statistical analysis
- Expression quantitative trait locus analyses
- Hardy–Weinberg equilibrium
- Meta analysis
Results
1. Within this study, researchers conducted a meta-analysis on 10,557 participants of European ancestry. The findings divulged that for every C allele copy, when adjusted for baseline HbA1c, there was a 0.07% reduction in glycated hemoglobin. Without adjustments, the allelic effect of the C allele was established as 0.17.
 Figure 1 Pharmacogenetic impact of rs8192675 on metformin response in participants of European ancestry.
Figure 1 Pharmacogenetic impact of rs8192675 on metformin response in participants of European ancestry.
2. To narrow down the list of potentially pathogenic genes and mutations, a localized meta-analysis was used in this research. The mutation type rs8192675 and its proxies demonstrated the strongest correlation with a reduction in HbA1c levels. Scientists assessed if rs8192675 acted as an expression quantitative trait locus (eQTL) in 1,226 liver samples with European ancestry. The results revealed that rs8192675, as the highest cis-eQTL of SLC2A2, demonstrated lowered expression of the 'C' allele. This suggested that the 'C' allele at the rs8192675 locus was significantly associated with decreased SLC2A2 expression.
 Figure 2 Regional plots of the SLC2A2 locus.
Figure 2 Regional plots of the SLC2A2 locus.
3. In this study, the investigators also explored the potential clinical implications of rs8192675. Their findings revealed that the clinical potential was more significant in obese patients, where homozygous carriers of the C allele experienced a 0.33% decrease in HbA1c percentages compared to homozygous carriers of the T allele.
Conclusion
This study established a robust multi-ethnic cohort relationship between rs8192675 and the reduction in HbA1c induced by Metformin. Metformin is shown to improve the defects in glucose homeostasis present in type 2 diabetes patients before treatment due to the reduced expression of SLC2A2. Moreover, in obese patients, rs8192675 could serve as a biomarker for stratified medicine.
Reference:
- Zhou K, Yee S W, Seiser E L, et al. Variation in the glucose transporter gene SLC2A2 is associated with glycemic response to metformin. Nature genetics, 2016, 48(9): 1055-1059.


 Sample Submission Guidelines
Sample Submission Guidelines