The Yeast DNA Kit was created specifically for isolating genomic DNA from yeast and other starting specimens. Within one hour, purified genomic DNA can be retrieved from yeast specimens. Its method of extraction does not include phenol or chloroform. Due to its unique silica membrane innovation, buffer solution, and other organic compounds, the Kit provides high purity and stable quality DNA free of proteins. Restriction analysis, PCR analysis, Southern Blot, and library construction can all be done directly with purified DNA. The sample size required is no more than 5X10^7 cells.
Species:
Yeast and other starting samples
Storage:
Store at room temperature (15-25℃)
Components:
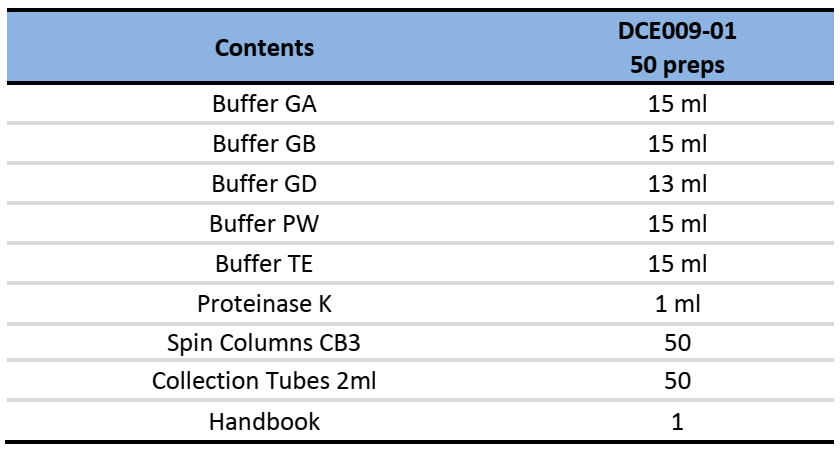
Specifications:
| Sample amount | no more than 5X107 cells |
| Features | Purified genomic DNA can be extracted from yeast samples within one hour. No phenol / chloroform extraction. Purified DNA can be used directly for PCR, restriction digestion and Southern Blot. |
| Application | The purified DNA can be used directly for molecular biological experiments such as restriction analysis, PCR analysis, Southern Blot and library construction. |
Ensure that ethanol (96-100%) has been added into Buffer GD and Buffer PW as indicated on the tag of the bottle before use.
- Pellet an appropriate amount of yeast cell (no more than 5 × 107 cells) in a microcentrifuge tube, centrifuge for 1 min at 12,000 rpm (~13,400 × g). Discard supernatant.
Note: If the sample is more than 700 μl, repeat the centrifugation process to collect all yeast cells into one tube. - Lysis (enzyme method) to crack yeast cell wall: Add 600 μl Sorbitol Buffer and 50 U Lyticase (should be prepared by user). Mix thoroughly, and incubate at 30°C for 30 min. Centrifuge for 10 min at 4,000 rpm (~1500 × g). Discard supernatant.
- Add 200 μl Buffer GA to resuspend the cell pellet. Mix thoroughly by vortex.
Note: If RNA-free genomic DNA is required, add 4 μl RNase A (100 mg/ml, should be prepared by user ). Mix by vortex for 15 s, and incubate for 5 min at room temperature (15-25°C). - Add 20 μl Proteinase K. Mix thoroughly.
- Add 220 μl Buffer GB to the sample, mix thoroughly by upsides and down, and incubate at 70°C for 10 min to yield a homogeneous solution. Briefly centrifuge the microcentrifuge tube to remove drops from the inside of the lid.
- Add 220 μl ethanol (96-100%) to the sample, and mix thoroughly by upsides and down. White precipitates may form on addition of ethanol. Centrifuge briefly to remove drops from the inside of the lid.
- Pipet the mixture from step 6 into the Spin Column CB3 (in a 2ml collection tube) and centrifuge at 12,000 rpm (~13,400 × g) for 30 s. Discard flow-through and place the spin column into the collection tube.
- Add 500 μl Buffer GD (Ensure ethanol has been added) to Spin Column CB3, and centrifuge at 12,000 rpm (~13,400 × g) for 30 s, then discard the flow-through and place the spin column back to the collection tube.
- Add 600 μl Buffer PW (Ensure ethanol has been added) to Spin Column CB3, and centrifuge at 12,000 rpm (~13,400 × g) for 30 s. Discard the flow-through and place the spin column back to the collection tube.
- Repeat step 9.
- Centrifuge at 12,000 rpm (~13,400 × g) for 2 min to dry the membrane completely. Open lid of CB3 and stay at room temperature for a while to dry the membrane completely.
- Place the Spin Column CB3 in a new clean 1.5 ml microcentrifuge tube, and pipet 50-200 μl Buffer TE or distilled water directly to the center of the membrane. Incubate at room temperature (15-25°C) for 2-5 min, and then centrifuge for 2 min at 12,000 rpm (~13,400 × g).
- (Optional) For increased DNA concentration, add the solution obtained from step 12 to the center of membrane again. Incubate at room temperature (15-25°C) for 2-5 min, and then centrifuge for 2 min at 12,000 rpm (~13,400 × g).



