CD RNA Adapters S3-S6 for Illumina are designed specifically for the preparation of CD RNA libraries on Illumina platforms. There are four kits in the kit and each kit includes a Universal Adapter, as well as 24 different 8bp, indexed RNA Adapters that can be used to differentiate between samples. It is a dedicated kit for Illumina high-throughput sequencing platform library construction, ideal for Illumina high-throughput sequencing platform multi-sample RNA library construction.
Components:
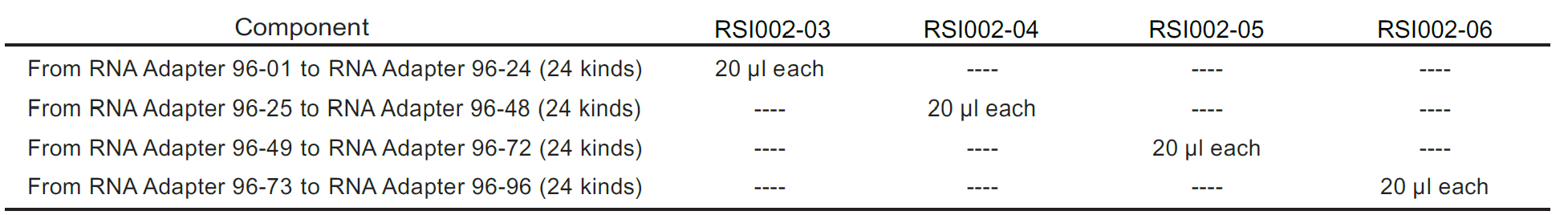
Application:
Special for RNA library preparation for Illumina platforms with CD mRNA-seq Library Prep Kit for Illumina (RP001), CD Stranded mRNA-seq Library Prep Kit for Illumina (RP002) or CD Total RNA-seq Library Prep Kit for Illumina(human/mouse/rat) (RP003).
Storage:
All the components can be stored at -20℃ for one year.
Specifications:
| Application | Used for: CD mRNA-seq Library Prep Kit for Illumina CD Stranded mRNA-seq Library Prep Kit for Illumina CD Total RNA-seq Library Prep Kit for Illumina(human/mouse/rat) |
| Sequencing Platform | Illumina |
The structure of libraries prepared with CD RNA Adapters s3-s6 for Illumina are as follows:
5’ - Universal Adapter - Insert RNA Sequence - DNA Adapter 96-X - 3’
Both the Universal Adapter and the Index are contained in each DNA Adapter. The sequences of RNA Adapters are as follows:
| Name | Sequence |
| Universal Adapter | 5’-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3’ |
| RNA Adapter 96-X | 5’-GATCGGAAGAGCACACGTCTGAACTCCAGTCACIIIIIIIIATCTCGTATGCCGTCTTCTGCTTG-3’ |
IIIIIIII indicates the index sequences (8 bp). Please input the related Index sequences of RNA Adapters to the Sample Sheet before sequencing as follows:
| RSI002-03 | Index | RSI002-04 | Index | RSI002-05 | Index | RSI002-06 | Index |
| RNA Adapter 96-01 | AACGTGAT | RNA Adapter 96-25 | AGATCGCA | RNA Adapter 96-49 | GATAGACA | RNA Adapter 96-73 | AATGTTGC |
| RNA Adapter 96-02 | AAACATCG | RNA Adapter 96-26 | AGCAGGAA | RNA Adapter 96-50 | GCCACATA | RNA Adapter 96-74 | ACACGACC |
| RNA Adapter 96-03 | ATGCCTAA | RNA Adapter 96-27 | AGTCACTA | RNA Adapter 96-51 | GCGAGTAA | RNA Adapter 96-75 | ACAGATTC |
| RNA Adapter 96-04 | AGTGGTCA | RNA Adapter 96-28 | ATCCT GTA | RNA Adapter 96-52 | GCTAACGA | RNA Adapter 96-76 | AGATGTAC |
| RNA Adapter 96-05 | ACCACTGT | RNA Adapter 96-29 | ATTGAGGA | RNA Adapter 96-53 | GCTCGGTA | RNA Adapter 96-77 | AGCACCTC |
| RNA Adapter 96-06 | ACATTGGC | RNA Adapter 96-30 | CAACCACA | RNA Adapter 96-54 | GGAGAACA | RNA Adapter 96-78 | AGCCATGC |
| RNA Adapter 96-07 | CAGATCTG | RNA Adapter 96-31 | GACTAGTA | RNA Adapter 96-55 | GGTGCGAA | RNA Adapter 96-79 | AGGCTAAC |
| RNA Adapter 96-08 | CATCAAGT | RNA Adapter 96-32 | CAATGGAA | RNA Adapter 96-56 | GTACGCAA | RNA Adapter 96-80 | ATAGCGAC |
| RNA Adapter 96-09 | CGCTGATC | RNA Adapter 96-33 | CACTTCGA | RNA Adapter 96-57 | GTCGTAGA | RNA Adapter 96-81 | ATCATTC C |
| RNA Adapter 96-10 | ACAAGCTA | RNA Adapter 96-34 | CAGCGTTA | RNA Adapter 96-58 | GTCTGTCA | RNA Adapter 96-82 | ATTGGCTC |
| RNA Adapter 96-11 | CTGTAGCC | RNA Adapter 96-35 | CATACCAA | RNA Adapter 96-59 | GTGTTCTA | RNA Adapter 96-83 | CAAGGAGC |
| RNA Adapter 96-12 | AGTACAAG | RNA Adapter 96-36 | CCAGTTCA | RNA Adapter 96-60 | TAGGATGA | RNA Adapter 96-84 | CACCTTAC |
| RNA Adapter 96-13 | AACAACCA | RNA Adapter 96-37 | CCGAAGTA | RNA Adapter 96-61 | TATCAGCA | RNA Adapter 96-85 | CCATCCTC |
| RNA Adapter 96-14 | AACCGAGA | RNA Adapter 96-38 | CCGTGAGA | RNA Adapter 96-62 | TCCGTCTA | RNA Adapter 96-86 | CCGACAAC |
| RNA Adapter 96-15 | AACGCTTA | RNA Adapter 96-39 | CCTCCTGA | RNA Adapter 96-63 | TCTTCACA | RNA Adapter 96-87 | CCTAATCC |
| RNA Adapter 96-16 | AAGACGGA | RNA Adapter 96-40 | CGAACTTA | RNA Adapter 96-64 | TGAAGAGA | RNA Adapter 96-88 | CCTCTATC |
| RNA Adapter 96-17 | AAGGTACA | RNA Adapter 96-41 | CGACTGGA | RNA Adapter 96-65 | TGGAACAA | RNA Adapter 96-89 | CGACACAC |
| RNA Adapter 96-18 | ACACAGAA | RNA Adapter 96-42 | CGCATACA | RNA Adapter 96-66 | TGGCTTCA | RNA Adapter 96-90 | CGGATTGC |
| RNA Adapter 96-19 | ACAGCAGA | RNA Adapter 96-43 | CTCAATGA | RNA Adapter 96-67 | TGGTGGTA | RNA Adapter 96-91 | CTAAGGTC |
| RNA Adapter 96-20 | ACCTCCAA | RNA Adapter 96-44 | CTGAGCCA | RNA Adapter 96-68 | TTCACGCA | RNA Adapter 96-92 | GAACAGGC |
| RNA Adapter 96-21 | ACGCTCGA | RNA Adapter 96-45 | CTGGCATA | RNA Adapter 96-69 | AACTCACC | RNA Adapter 96-93 | GACAGTGC |
| RNA Adapter 96-22 | ACGTATCA | RNA Adapter 96-46 | GAATCTGA | RNA Adapter 96-70 | AAGAGATC | RNA Adapter 96-94 | GAGTTAGC |
| RNA Adapter 96-23 | ACTATGCA | RNA Adapter 96-47 | CAAGACTA | RNA Adapter 96-71 | AAGGACAC | RNA Adapter 96-95 | GATGAATC |
| RNA Adapter 96-24 | AGAGTCAA | RNA Adapter 96-48 | GAGCTGAA | RNA Adapter 96-72 | AATCCGTC | RNA Adapter 96-96 | GCCAAGAC |
Quality Control
16-Hour Incubation: A 50 ul reaction system containing 5 ul of Oligos and 1 μg of Hind III-λ DNA incubated at 37 °C for 16 hours resulted in no band degraded detected by agarose gel electrophoresis. A 50 μl reaction system containing 5 ul of Oligos and 1 μg of T3 DNA incubated at 37 °C for 16 hours resulted in no band degraded detected by agarose gel electrophoresis.
Endonuclease Activity: A 50 ul reaction system containing 5 ul of RNA adapters and 1 ug of pX174RF I DNA incubated at 37°C for 4 hours resulted in < 10% conversion to RF Il analyzed by agarose gel electrophoresis.



