The CD RRBS Library Kit simplifies the process flow for next-generation sequencing (NGS) applications on Illumina platforms using the Reduced Representation Bisulfite Sequencing (RRBS) technique. To enhance for CpG dinucleotides, RRBS (Reduced Representation Bisulfite Sequencing) integrates restriction enzyme digestion with bisulfite sequencing. RRBS lowers the complexity of the genome and the cost of sequencing by capturing a large amount of methylation data. This kit facilitates library preparation with good purity and integrity from as little as 10ng genomic DNA. Its workflow is quick and easy. It also ensures that library preparation is consistent and repeatable. This kit is preferred for: Promoter and CpG islands coverage analysis Promoter and CpG island methylation level analysis DMR (differentially methylated area) analysis, Functional annotation of differentially expressed genes, Relation of methylation level and functional elements’ length, CpG area distribution and Regional distribution of genes.
Storage:
Box 1 should be stored at room temperature (20-30°C). Box 2 should be stored at -20℃
Components:
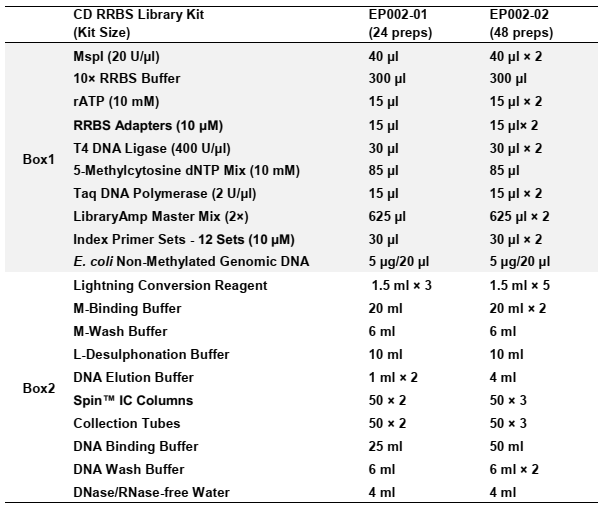
Specifications:
| Sample amount | 10ng-500ng |
| Features | Fast and convenient workflow. Ensures the consistency and reproducibility of library preparation. |
| Application | It is recommended to use this kit for: Promoter and CpG islands coverage analysis Promoter and CpG island methylation level analysis DMR (differentially methylated region) analysis Functional annotation of differentially expressed genes Relation of methylation level and functional elements’ length CpG region distribution Regional distribution of genes |
| Species Category | mammals |
| Sample type | Purified genomic DNA free of enzymatic inhibitors |
| Sequencing Platform | Illumina |
MspI Digestion
- Mix 10× RRBS Buffer, MspI, Genomic DNA in TE buffer/H2O, and DNase/RNase-free Water in a 0.2 ml PCR tube.
- Incubate the mixture in a thermocycler.
Adapter Ligation
- Add 10× RRBS Buffer, rATP, RRBS Adapters, MspI, T4 DNA Ligase, and DNase/RNase-free Water to the Mspl Digestion step for adapter ligation.
- Mix well and incubate the mixture in a thermocycler.
- Add Taq DNA Polymerase, and 5-methylcytosine dNTP Mix to the ligation product.
- Mix well and incubate the mixture in a thermocycler at 74°C for 30 minutes.
- In a 1.5 ml tube, add a 7:1 volume ratio of DNA Binding Buffer to the product from step 4, mix well, and transfer to a Spin™ IC column in a Collection Tube. Centrifuge at ≥ 10,000 x g for 30 seconds.
- Add 200 μl DNA Wash Buffer to the column. Centrifuge at ≥ 10,000 x g for 30 seconds. Discard the flow-through. Repeat this wash step.1
- Transfer the Spin™ IC column to a 1.5 ml tube. Add 20 μl DNA Elution Buffer directly to the column matrix and let stand for 1 minute at room temperature2. Centrifuge at ≥ 10,000 x g for 30 seconds to elute.
Bisulfite Conversion
- Mix Lightning Conversion Reagent, and product from the Adapter Ligation step in a 0.2 ml PCR tube.
- Incubate the mixture in a thermocycler.
- Add 600 μl M-Binding Buffer to a Spin™ IC Column in a Collection Tube. Add the bisulfite converted sample to the column, close the cap, and invert ~ 8 times to mix. Centrifuge at ≥ 10,000 x g for 30 seconds.
- Discard the flow-through from the Collection Tube and add 100 μl M-Wash Buffer to the column. Centrifuge at ≥ 10,000 x g for 30 seconds.
- Add 200 μl L-Desulphonation Buffer to the column and let stand at room temperature (20°C-30°C) for 15-20 minutes.2 After the incubation, centrifuge at ≥ 10,000 x g for 30 seconds.
- Add 200 μl M-Wash Buffer to the column and centrifuge at ≥ 10,000 x g for 30 seconds. Discard the flow-through. Repeat this wash step.
- Transfer the Spin™ IC column to a 1.5 ml tube. Add 24 μl DNA Elution Buffer directly to the column matrix and let stand for 1 minute. Centrifuge at ≥ 10,000 x g for 30 seconds to elute the bisulfite-converted DNA.
Index Primer Amplification
- Mix LibraryAmp Master Mix, Index Primer Set, and product from the Bisulfite Conversion step in a 0.2 ml PCR tube.
- Incubate the mixture in a thermocycler to amplify the libraries.
- Purify the PCR product following steps 5 to 7 from Adapter Ligation and elute in 15 μl DNA Elution Buffer. If using agarose gel to validate the size distribution of the libraries, either go to step 1 in Section 5 prior to PCR clean-up or elute in a higher volume to ensure the volumes of libraries are large enough for both validation and quantification.
Library Validation and Quantification
- The size distribution of the libraries from Section 4 can be visualized using gel electrophoresis. No primer dimer bands should appear at ~ 100 – 120 bp.
- CD Library Quantification Kit for Illumina (LQ001) or similar for Illumina® platforms are recommended for library quantification prior to sequencing. The libraries are ready for sequencing after CD Library quantification.
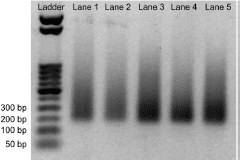
The size distribution of the libraries from Index Primer Amplification



