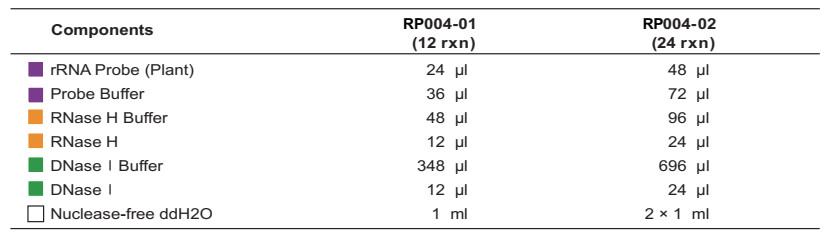
The rRNA Depletion Kit (Plant) is intended to remove ribosomal RNA (rRNA) from total RNA, such as cytoplasmic rRNA (5S, 18S, and 25S rRNA), mitochondrial rRNA, and chloroplast rRNA, from a variety of sample types, such as plant roots, seeds, and leaves. This kit allows you to remove rRNA from 1g to 5g of intact or degraded total RNA. The kit includes rRNA hybridization probe, RNase H digestion, DNaseI digestion, buffers, and other reagents required for rRNA depletion. Following the digestion steps, the retained mRNA and other non-coding RNA can be used to create an RNA library that can be used to analyze LncRNA and other non-coding RNAs.
Storage:
All the components should be stored at -20℃.
Specifications:
| Sample amount | 1µg -5µg |
| Features | Compatible with a wide range of sample types, including plant roots, seeds and leaves from different plant species. Remove cytoplasmic rRNA (5S, 18S and 25S rRNA), mitochondrial rRNA and chloroplast rRNA effectively. |
| Application | The obtained RNA can be used for transcriptome library construction with strand-specific information or non-strand-specific information. |
| Species Category | compatible with a wide range of sample types, including plant roots, seeds and leaves |
| Sample type | intact or degraded total RNA |
| Sequencing Platform | Illumina |
Workflow
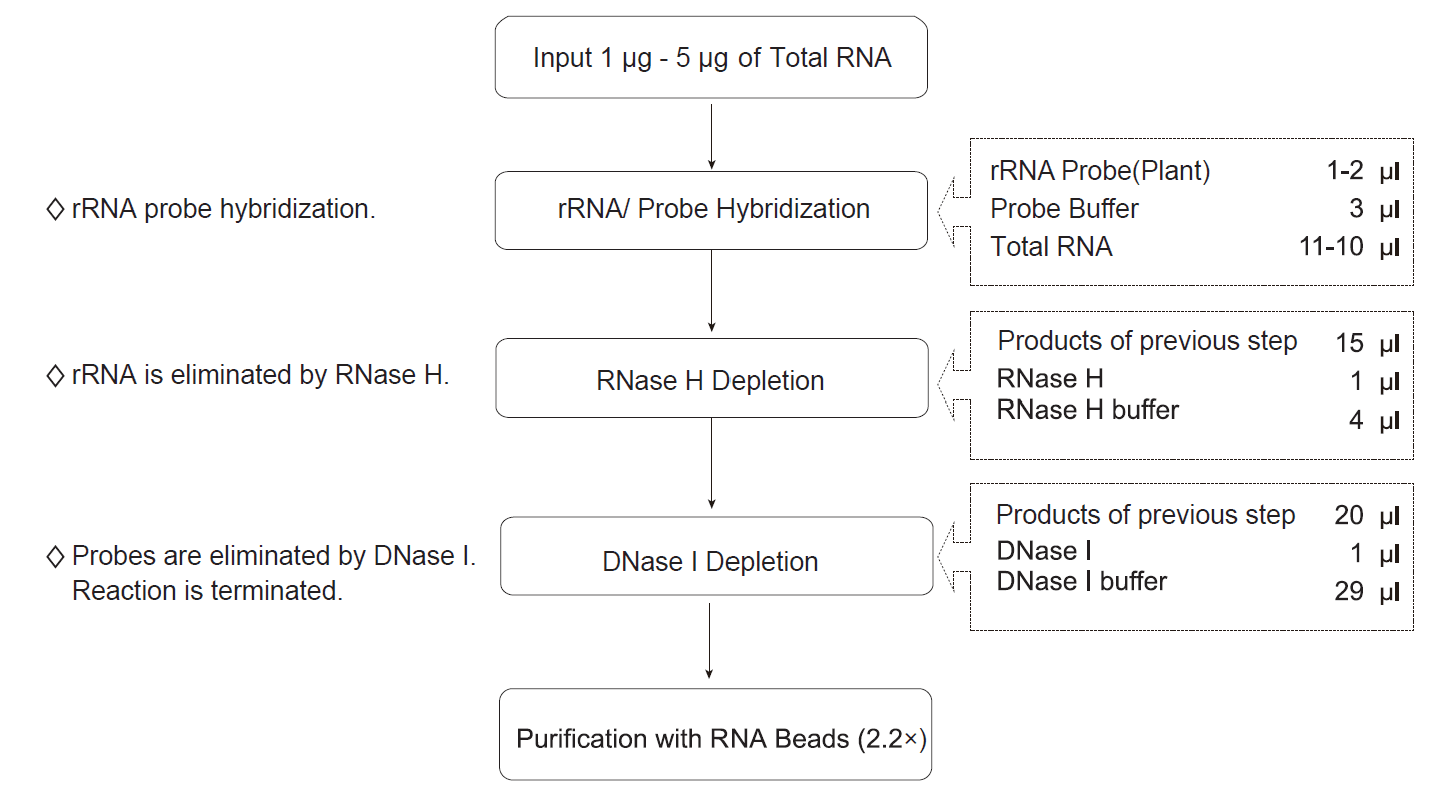 Workflow of CD rRNA Depletion Kit (Plant)
Workflow of CD rRNA Depletion Kit (Plant)
Protocol
rRNA/Probe hybridization
- Prepare total RNA samples: Dilute 1 μg - 5 μg of total RNA with 11 μl or 10 μl of Nuclease-free Water in a Nuclease-free PCR tube and keep the tube on ice for use.
- Prepare the reaction solution in a Nuclease-free centrifuge tube.
- Put the sample into a PCR instrument and run the program.
Digestion with RNase H
Prepare the digestion with DNase H reaction solution on ice. Mix thoroughly by gently pipetting up and down for 10 times. Collect the liquid to the bottom of the tube by a brief centrifugation. Then put the sample into a PCR instrument and run the program.
Digestion with DNase I
Prepare the digestion with DNase I reaction solution on ice. Mix thoroughly by gently pipetting up and down for 10 times. Collect the liquid to the bottom of the tube by a brief centrifugation. Then put the sample into a PCR instrument and run the program.
Purification of Ribosomal-depleted RNA with CD RNA Clean Beads
- Suspend the CD RNA Clean Beads thoroughly by vortexing, pipet 110 µl (2.2 ×) of beads into the RNA sample of Step 08-3. Mix thoroughly by pipetting up and down for 10 times.
- Incubate the sample on ice for 15 min to make the RNA bind to the beads.
- Put the sample onto a magnetic stand. Wait until the solution clarifies (about 5 min). Then carefully discard the supernatant without disturbing the beads.
- Keep the sample on the magnetic stand, add 200 µl of 80% Ethanol (freshly prepared using Nuclease-free ddH2O) to rinse the beads. Incubate at room temperature for 30 sec and carefully discard the supernatant without disturbing the beads.
- Repeat Step 4.
- Keep the sample on the magnetic stand, open the tube and air-dry the beads for 5 min - 10 min.
- Option A (if the ribosomal-depleted RNA will be used for reverse transcription): take the sample out of magnetic stand, add 20 µl of Nuclease-free ddH2O and mix thoroughly by pipetting for 6 times, and incubate at room temperature without shaking for 2 min. Put the tube back on the magnetic stand and wait until the solution clarifies (about 5 min), carefully transfer 18 µl of the supernatant to a new Nuclease-free PCR tube without disturbing the beads.
Option B (if the ribosomal-depleted RNA will be used for RNA library preparation with CD Stranded mRNA-seq Library Prep Kit for Illumina) (RP002): take the sample out of magnetic stand, add 18.5 µl of Frag/Primer Buffer and mix thoroughly by pipetting up and down for 6 times, and incubate at room temperature without shaking for 2 min. Put the tube back on the magnetic stand and wait until the solution clarifies (about 5 min), carefully transfer 16 µl of the supernatant to a new Nuclease-free PCR tube without disturbing the beads for library preparation.