The total RNA-seq Library Prep Kit for Illumina (human/mouse/rat) offers a quick and easy workflow for strand-specific library preparation in Illumina platforms for next-generation sequencing (NGS). This kit allows you to make libraries from as little as 100ng to 1g total RNA from human, mouse, or rat specimens, and it's consistent with a wide range of sample types, such as low-quality RNA and formalin-fixed, paraffin-embedded (FFPE) samples. This kit includes all necessary reagents for the removal of ribosomal RNA (rRNA), including both cytoplasmic (28S, 18S, 5.8S, and 5S) and mitochondrial (16S and 12S) rRNA, in comparison to traditional transcriptome library preparation kits. Not only does the prepared library contain mRNA, but it also contains multiple types of non-coding RNA information. As a result, the Kit is appropriate for non-coding RNA analysis, such as lncRNA. During the second strand synthesis, actinomycin D is used to maintain strand specificity, and dUTP is used to ensure stranded library construction. Before library enrichment, the second strand containing dUTP will be digested with uracil-DNA glycosylase (UDG). As a result, only the information from the first strand of cDNA is kept. All Kit components are subjected to rigorous quality control to ensure that library preparation is consistent and repeatable.
Storage:
All the components should be stored at -20℃.
Components:
-Components.png)
Specifications:
| Sample amount | 100ng-1µg |
| Features | The Ribo-Off rRNA depletion module contains all necessary reagents for the removal of ribosomal RNA (rRNA) including both cytoplasmic (28S, 18S, 5.8S, and 5S) and mitochondrial (16S and 12S) rRNA. Obtained strand-specific information is helpful to bioinformatics analysis. |
| Application | It is recommended to use this kit for: Gene expression analysis Single nucleotide variation calling Gene fusion detection Transcript Variants Novel Transcripts Target transcriptome analysis |
| Species Category | human, mouse, rat |
| Sample type | intact or degraded total RNA, compatible with FFPE samples |
| Sequencing Platform | Illumina |
Workflow
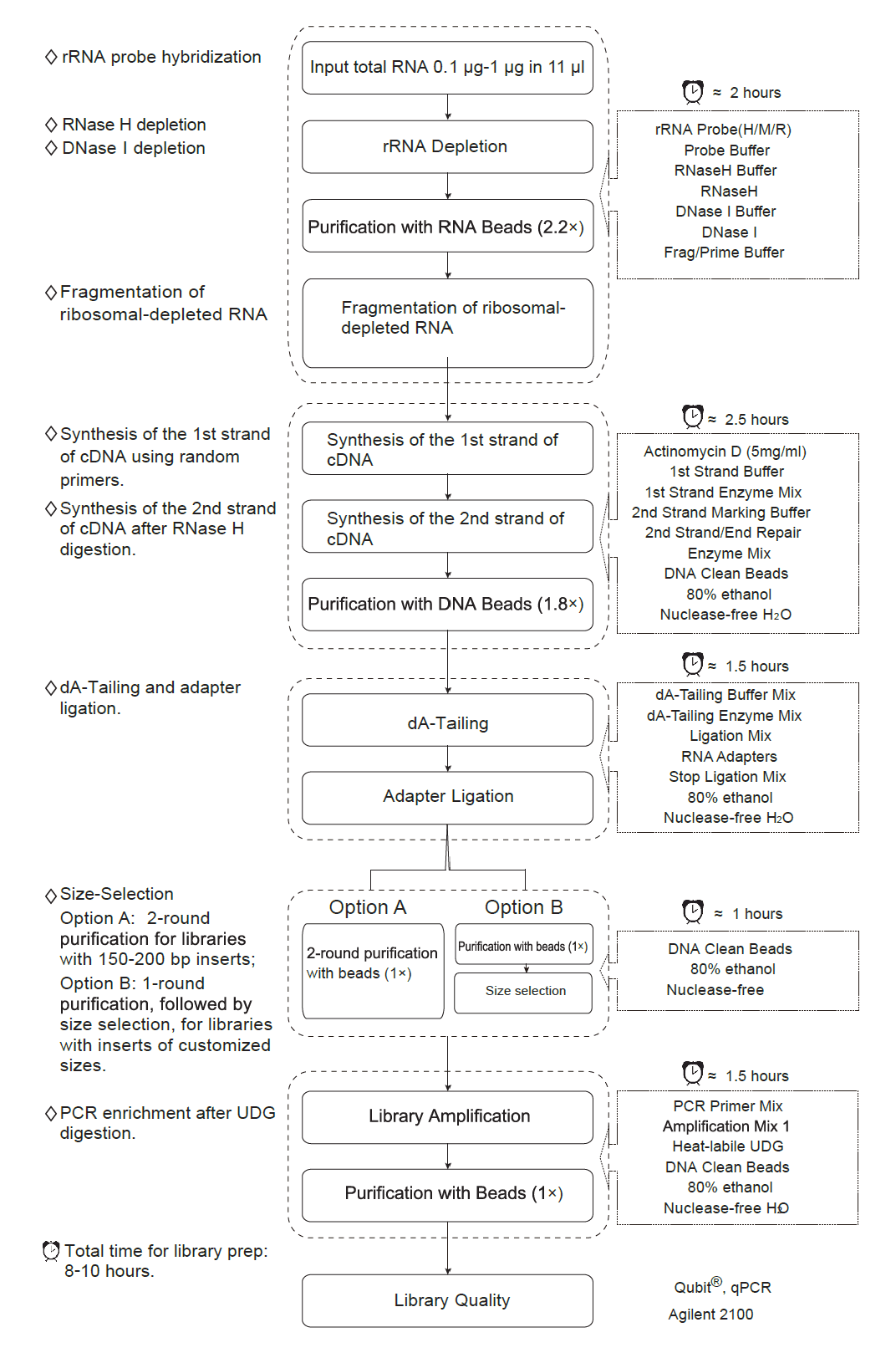 Workflow of CD Total RNA-seq Library Prep Kit for Illumina (human/mouse/rat)
Workflow of CD Total RNA-seq Library Prep Kit for Illumina (human/mouse/rat)
Protocol
rRNA Depletion and Fragmentation
- Preparation of total RNA sample: Dilute 0.1 µg - 1 µg of total RNA with 11 µl of Nuclease-free Water in a Nuclease-free PCR tube and keep on ice.
- rRNA/probe hybridization.
- Digestion with RNase H.
- Digestion with DNase I.
- Purification of ribosomal-depleted RNA with CD RNA Clean Beads
Synthesis of Double Strand cDNA
- Tap the tube several times to mix thoroughly, collect the liquid at the bottom of the tube by a brief centrifugation. Dilute to 0.5 mg/ml by adding 1 µl Actinomycin D (5 mg/ml) to 9 µl Nuclease-free Water and use immediately.
- Thaw the 1st Strand Buffer and mix it thoroughly by inverting the tube. Prepare the reaction solution to synthesize the first strand of cDNA.
- Put the sample in a PCR instrument and run PCR program.
- Thaw the 2nd Strand Buffer and mix it thoroughly by inverting the tube. Prepare the reaction solution to synthesize the 2nd strand of cDNA.
- Put the sample in a PCR instrument and run an incubation program.
- Purification of the double strand cDNA.
Note: The Diluted Actinomycin D solution is highly sensitive to light and will attach to the surfaces of plastic and glass. Discard unused diluted solution.
dA-Tailing
- Thaw the dA-tailing Buffer Mix and mix it thoroughly by inverting the tube. Prepare the reaction solution.
- Put the sample in a PCR instrument and run PCR program for dA-Tailing.
Adapter Ligation
- Thaw the RNA Adapter and mix it thoroughly by inverting the tube. Prepare the reaction solution.
- Put the sample in a PCR instrument and run program for adapter ligation.
- Add 5 µl of the Stop Ligation Mix to 35 µl of ligation products, mix thoroughly by gently pipetting up and down for 10 times to terminate the ligation reaction.
Purification and Size Selection of Adapter-ligated DNA
There are two options in this step, please choose according to the length of the inserts.
Option A for libraries with 150-200 bp inserts (for mRNA fragmented by incubation at 94℃ for 8 min).
Option B for libraries with > 200 bp inserts (for mRNA fragmented by incubation at 94℃ for 5 min, 85℃ for 6 min, or 85℃ for 5 min).
Library Amplification
- Thaw the PCR Primer Mix and Amplification Mix 1 thoroughly by inverting the tube. Prepare the reaction solution.
- Put the sample in a PCR instrument and run the PCR program.
- Purification of the PCR product with CD DNA Clean Beads.
- Library Quality Determination.


