The Stranded mRNA-seq Library Prep Kit for Illumina offers a quick and easy workflow for library preparation in Illumina systems' next-generation sequencing (NGS) applications. This kit allows library preparation from as little as 50ng to 4g total RNA from animals, plants, fungi, and other eukaryotes with high purity and integrity. This Kit, unlike non-stranded transcriptome library prep kits, uses actinomycin D to maintain strand specificity and dUTP to ensure stranded library construction during the second strand synthesis. Before library enrichment, the dUTP-containing second strand will be digested with uracil-DNA glycosylase (UDG). As a result, only the information from the first strand of cDNA is saved. All Kit components go through a rigorous quality control process to ensure that library preparation is consistent and repeatable.
Storage:
Box 1 should be stored at 2℃ - 8℃. Box 2 should be stored at -30℃ ~ -15℃
Components:
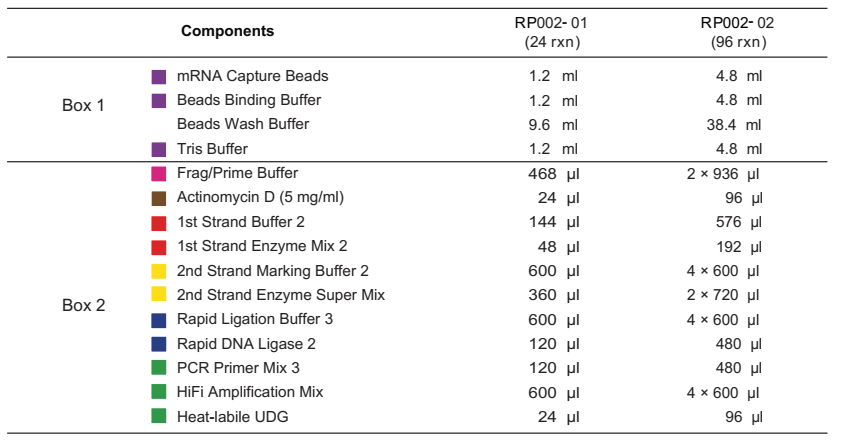
Specifications:
| Sample amount | 50ng-4µg |
| Features | Fast and convenient workflow enables a single library preparation within one day. Captures the coding transcriptome with strand information. |
| Application | It is recommended to use this kit for: Gene expression analysis Single nucleotide variation calling Gene fusion detection Transcript Variants Novel Transcripts Target transcriptome analysis |
| Species Category | animals, plants, fungus, and other eukaryotes |
| Sample type | total RNA with good purity and integrity |
| Sequencing Platform | Illumina |
Workflow
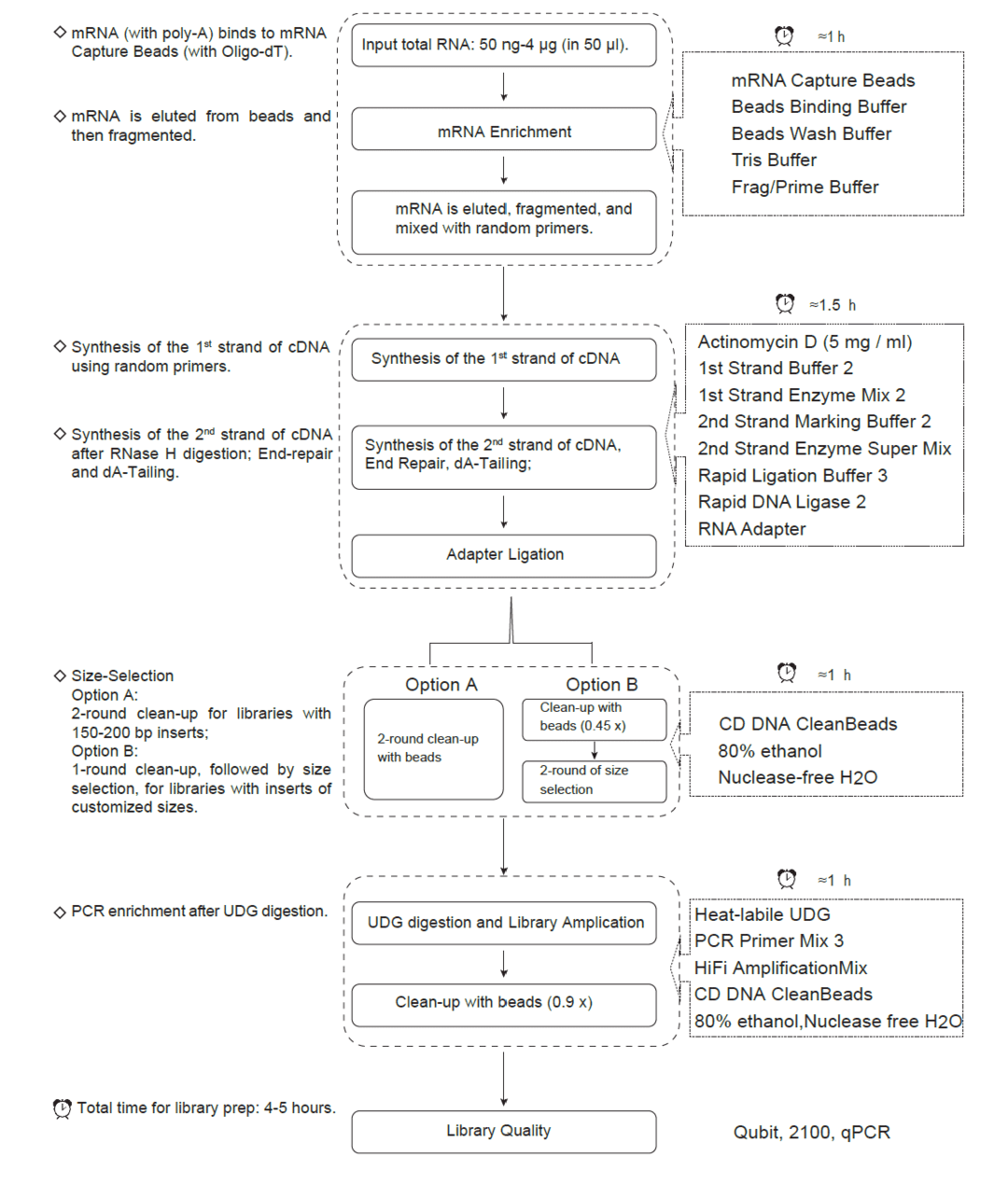 Workflow of CD Stranded mRNA-seq Library Prep Kit for Illumina ®
Workflow of CD Stranded mRNA-seq Library Prep Kit for Illumina ®
Protocol
mRNA Purification and Fragmentation
- Equilibrate the BOX 1 to room temperature, which contains mRNA Capture Beads, Beads Wash Buffer, Tris Buffer, and Beads Binding Buffer.
- Prepare the reaction solution.
- Run PCR program for the binding of mRNA to mRNA-Capture Beads.
- Put the samples onto a magnetic stand. Wait until the solution clarifies (about 5 min), then carefully discard the supernatant without disturbing the beads.
- Take the samples out of the magnetic stand. Add 200 µl of Beads Wash Buffer, and mix thoroughly by pipetting up and down for 10 times. Put the samples back to the magnetic stand. Wait until the solution clarifies (about 5 min), then carefully discard the supernatant without disturbing the beads.
- Take the samples out of the magnetic stand, and add 50 µl of Tris Buffer to re-suspend the beads thoroughly by pipetting up and down for 10 times.
- Run the following procedure in the PCR instrument to release mRNA.
- Add 50 µl of Beads Binding Buffer, mix thoroughly by pipetting up and down for 10 times.
- Incubate at room temperature for 5 min to make the mRNA bind to the beads.
- Place the samples on the magnetic stand to isolate the mRNA from total RNA. Wait until the solution clarifies (about 5 min), then carefully discard the supernatant without disturbing the mRNA Capture Beads.
- Take the samples out of the magnetic stand, add 200 µl of Beads Wash Buffer, and mix thoroughly by pipetting up and down for 10 times. Place the tube on the magnetic stand. Wait until the solution clarifies (about 5 min), then carefully discard the supernatant without disturbing the mRNA Capture Beads.
- Take the samples out of the magnetic stand, add 18.5 µl of Frag/Prime Buffer to re-suspend the beads thoroughly by pipetting up and down for 10 times. Incubate the samples in a PCR device and set programs according to fragment sizes:
- Place the samples on the magnetic stand. Wait until the solution clarifies (about 5 min), and pipet 16 µl of supernatant into a new Nuclease-free PCR tube, then immediately proceed to synthesis of 1st Strand cDNA.
Synthesis of Double Strand cDNA
The components for synthesis of double-stranded cDNA should be dissolved on ice, mixed upside down, briefly centrifuged to the bottom of the tube, and placed on ice for use.
- Dilute Actinomycin D (5 mg / ml) to 120 ng / μl.
- Prepare the reaction solution to synthesize the 1st strand of cDNA.
- Adjust the pipettor to a 20 μl range and mix thoroughly by gently pipetting up and down for 10 times.
- Run PCR program for the synthesis of 1st strand cDNA and 2nd strand cDNA.
Adapter Ligation
- Prepare the reaction solution of Adapter Ligation.
- Adjust the pipettor to an 80 μl range and mix thoroughly by gently pipetting up and down for 10 times.
- Run the program of ligation reaction in the PCR instrument.
Clean-up and Size Selection of Adapter-Ligated DNA
There are two options in this step, please choose carefully to follow as needed:
Option A: provides a 2-round clean-up without size-selection, and will generate fragments with 150 bp - 200 bp inserts (without residual adapters).
Option B: provides a 1-round clean-up and then a 2-round size-selection. According to different size-selection conditions in Table 2 (on page 7), this option will generate size-selected fragments with > 200 bp inserts (without residual adapters).
Library Amplification
- Prepare the PCR reaction system.
- Adjust the pipettor to a 30 μl range and mix thoroughly by gently pipetting up and down for 10 times.
- Run PCR program.
- Clean-up of the PCR product with CD DNA Clean Beads.
- Library Quality Analysis.


