The NEXT DNA Library Prep Kit for Illumina offers a quick and easy workflow for library preparation in Illumina-based next-generation sequencing (NGS) applications. The Kit incorporates the complex steps of DNA fragmentation, end-repair, dA-tailing, and adapter ligation into a one-step enzymatic reaction using a new-type transposase, which substantially decreases the quantity of initial DNA, lessening experiment time, and reducing the risk of contamination. This kit creates DNA-seq libraries from small amounts of DNA from a variety of sources and species, with three different specifications for different amounts of input DNA: 50ng, 5ng, and 1ng. All Kit components are subjected to rigorous quality control to ensure that library preparation is consistent and repeatable.
Storage:
TD501: can be stored at -20℃ for one year. TD502: 5× TS should be stored at room temperature(15-25℃); other components can be stored at -20℃ for one year. TD503: 5× TS should be stored at room temperature(15-25℃); other components can be stored at -20℃ for one year.
Components:
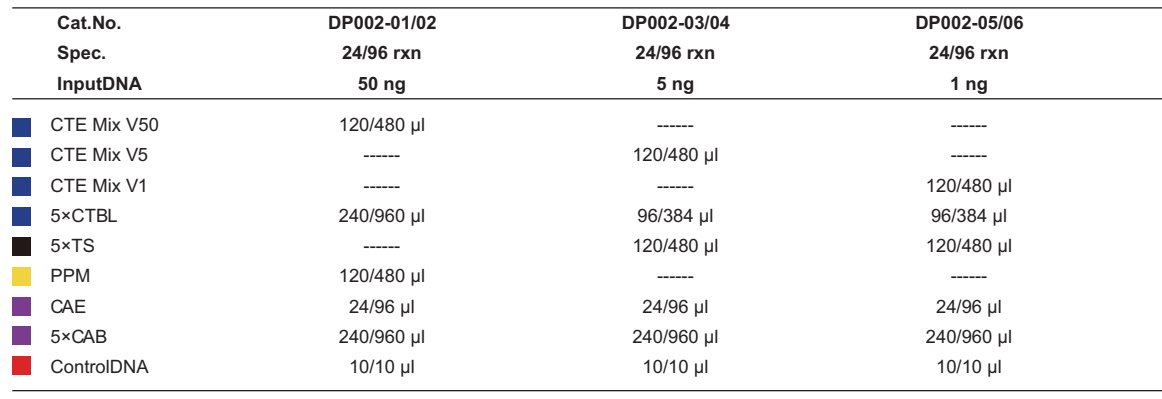
Specifications:
| Sample amount | 50ng,5ng,1ng |
| Features | Fast and convenient workflow enables a single library preparation within 90min. Based on a new-type transposase and special buffer system ensure efficient and uniform amplification. Suits to as low as 1ng amount of starting materials and provides three kinds of specifications for different amount of input DNA. |
| Application | It is recommended to use this kit for: Whole genome sequencing Whole exome or targeted sequencing (using Roche NimbleGen TM SeqCapTM EZ, Agilent SureSelect, Illumina TruSeq, or IDT xGen TM Lockdown TM Probes or other hybridization capture systems) Amplicon sequencing |
| Species Category | Compatible with any species |
| Sample type | genomic DNA and Amplicons, not FFPE-Compatible. It should be noted that the transposase in the Kit is not effective at the ends of DNA, leading to inevitable lower coverage of the 50bp on each end. Therefore, to avoid the reduction of sequencing coverage on the ends, it is recommended to extend 50-100bp at both ends of the to-be-sequenced region when PCR products are prepared. |
| Sequencing Platform | Illumina |
Workflow
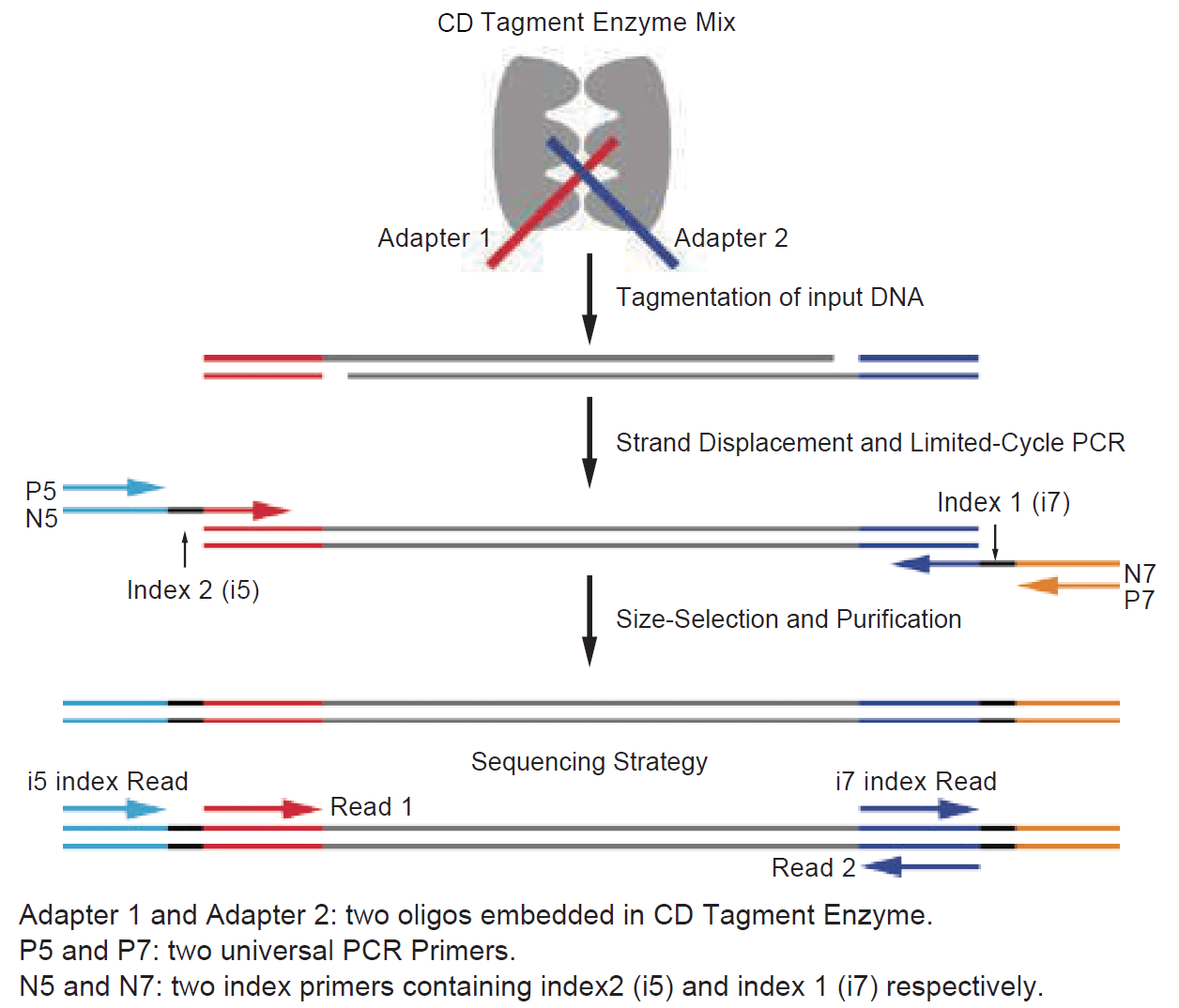 Workflow of CD NEXT DNA Library Prep Kit for Illumina ®
Workflow of CD NEXT DNA Library Prep Kit for Illumina ®
Protocol
Starting Material: Input DNA dissolved in sterile distilled water.
Determination of DNA Concentration: Since CTE Mix is sensitive to the concentration of DNA, the accuracy of DNA concentration determination is important to the success of the experiment. It is recommended to take advantage of Qubit® or PicoGreen® to determine the concentrations of DNA. Please avoid using methods based on the absorbance.
Requirement for DNA Purity: The absorbance of 260 nm / 280 nm = 1.8 - 2.0.
- DNA Tagmentation (select according to the kit Cat. No.)
- PCR Enrichment
2.1 Put the sterile PCR tube in the ice bath and add the components in order.
2.2 Gently pipet up and down to mix thoroughly. Put the PCR tube into a PCR instrument and run the program.
2.3 Proceed to Step 3. Purify the amplified library with CD DNA Clean Beads for size selection. - Purify the amplified library with CD DNA Clean Beads
- Library Quality Control
4.1 Library Concentration
In order to obtain a high quality of sequencing data, it is necessary to determine the accuracy concentration of the library. Real-time PCR was recommended to definitely quantify the concentration of the library. Besides, fluorescent dye methods (such as Qubit® and PicoGreen®) based on special recognition on double stranded DNA can also be utilized.
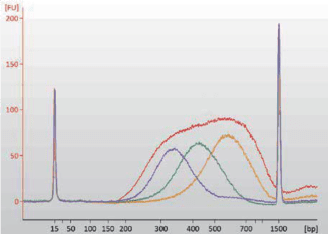
4.2 Library Distribution
Human genomic library prepared with CD NEXT DNA Library Prep Kit for Illumina (DP002-01, 9cycles of PCR Amplification) was analyzed by an Agilent Technologies 2100 Bioanalyzer.
It is recommended to use CD DNA Clean Beads to select and purify the amplified library. Equilibrate the beads to room temperature before use.
Before the next step, make up the volume to 50 μl with distilled water, for the product volume would be less than 50 μl due to the evaporation during PCR. It is important to make sure that the total volume is 50 μl, otherwise unexpected selected size may be obtained.
Tips
- CD DNA Clean Beads Tips
Equilibrate the beads to room temperature before use.
Mix the beads thoroughly every time before pipetting.
Thoroughly mix the beads with DNA samples.
All the DNA size selection and procedures using beads should be performed at room temperature. Do not pipet any CD DNA Clean Beads when transferring the supernatant.
Prepare fresh 80% ethanol and discard after use.
Try to remove all the 80% ethanol after washing.
Thoroughly air-dry the beads before DNA elution. - Avoid cross contamination between samples.
Change tips between samples.
Use filtered pipette tips. - Aliquot reagents after the first use to avoid repeated freeze-thaw cycles.
- Prevent contamination of PCR products.
Isolate the experimental area and carefully clean all equipments and instruments (e.g. clean with 0.5% sodium hypochlorite or 10% bleach) to avoid possible contamination in PCR system.



