The Rapid DNA Purification Kit is created to remove PCR products and DNA fragments from enzymatic reaction solutions quickly and easily. The Kit gives high purity DNA free of proteins, salts, and small fragment impurities, as well as >80% recovery yield for 100 bp -10 kb DNA fragments, thanks to its unique silica-membrane technology and buffer solution. Increased absorption and elution time could improve recovery efficiency for DNA fragments smaller than 100 bp and larger than 10 kb. Purified DNA can be directly used in molecular biological experiments like PCR, restriction enzyme digestion and hybridization, sequencing, and transformation, among others. It's worth noting that the Kit can be kept at room temperature (15-25°C) and that the amount of sample required is around 1-5 grams.
Storage:
Store at room temperature (15-25℃)
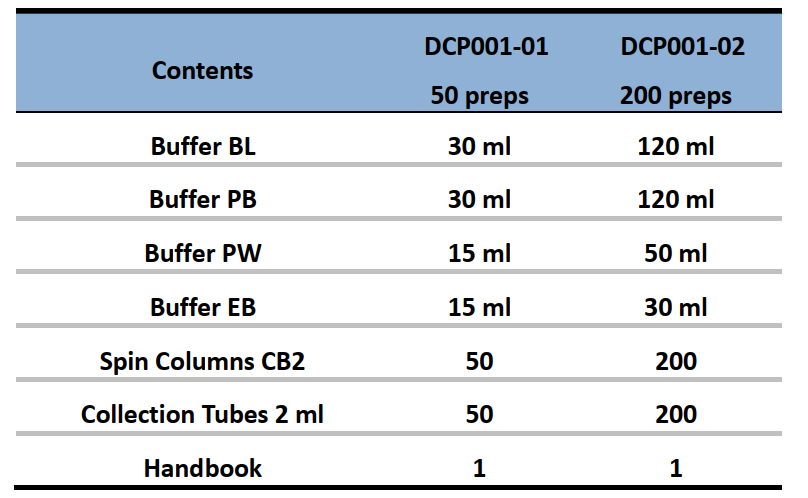
Components:
Specifications:
| Sample amount | 1-5µg |
| Features | Fast and convenient workflow enables DNA purification within 15min. Provides high purity DNA and >80% recovery yield for 100 bp -10 kb DNA fragments. 10 μg DNA binding capacity for each spin column. |
| Application | Suits to purify 100 bp-10 kb DNA fragments. The purified DNA can be used directly for molecular biological experiments such as PCR, restriction enzyme digestion and hybridization, sequencing, and transformation, etc. |
| Sample type | Isolate pure DNA from PCR products and DNA fragments from enzymatic reaction solutions. |
Add ethanol (96-100%) to Buffer PW before use (see bottle label for volume).
- Column equilibration: add 500 µl Buffer BL to the Spin Column CB2 (put Spin Column CB2 into a collection tube). Centrifuge for 1 min at 12,000 rpm (~13,400 × g). Discard the flow-through, and then place Spin Column CB2 back into the collection tube (please use freshly treated spin column).
- Add 5 volumes of Buffer PB to 1 volume of the PCR reaction or enzymatic reaction and mix. It is not necessary to remove mineral oil or kerosene.
- Transfer the mixture to the Spin Column CB2, incubate at room temperature (15-25°C) for 2 min. Centrifuge for 30-60 s at 12,000 rpm (~13,400 × g) in a table-top microcentrifuge. Discard the flow-through, and then place Spin Column CB2 back into the same collection tube.
- Add 600 µl Buffer PW (ensure that ethanol (96-100%) has been added) to the Spin Column CB2 and centrifuge for 30-60 s at 12,000 rpm (~13,400 × g). Discard the flow-through, and place Spin Column CB2 back in the same collection tube.
- Repeat step 4.
- Centrifuge at 12,000 rpm (~13,400 × g) for 2 min to remove residual Buffer PW. Discard the flow-through, and allow the column to air dry with the cap open for several minutes to dry the membrane.
- Place the Spin Column CB2 in a clean 1.5 ml microcentrifuge tube. Add 30-50 μl Buffer EB to the center of membrane, incubate for 2 min, and centrifuge for 2 min at 12,000 rpm (~13,400 × g).
Note: For example, add 250 μl Buffer PB to 50 μl PCR reaction (not including oil).
Note: The maximum loading volume of the column is 800 μl. For sample volumes greater than 800 μl simply load again.
Note: If the purified DNA is used for the subsequent salt sensitive experiments, such as ligation or sequencing experiment, it is suggested to stand for 2-5 min after adding Buffer PW, and then centrifuge.
Note: Residual ethanol from Buffer PW may inhibit subsequent experiment (enzymatic or PCR reactions).
Note: If the volume of eluted buffer is less than 30 μl, it may affect recovery efficiency. The pH value of eluted buffer will have big influence in eluting; distilled water (pH 7.0-8.5, adjusted with NaOH) is suggested to elute plasmid DNA, pH<7.0 will decrease elution efficiency. For long-term storage of DNA, eluting in Buffer EB and storing at -20°C is recommended, since DNA stored in water is subject to acid hydrolysis. Repeat step 7 to increase plasmid recovery efficiency.



