The FFPE DNA Kit was created specifically for isolating genomic DNA from formalin-fixed, paraffin-embedded tissues and is appropriate for a variety of samples. The Kit overpowers the inhibitory effect of formalin by using a unique deparaffinization solution and a specific lysis condition to release DNA. Furthermore, the one-of-a-kind deparaffinization solution is safer than xylene. The Kit delivers high-purity and high-integrity DNA in a small elution volume thanks to its unique silica-membrane technology and special buffer solution. Purified DNA can be used right away in molecular biology experiments like PCR, restriction enzyme digestion, and Southern hybridization. Roughly 5-8 pieces of 5-10 m of the sample are required.
Storage:
Store at room temperature (15-25℃)
Components:
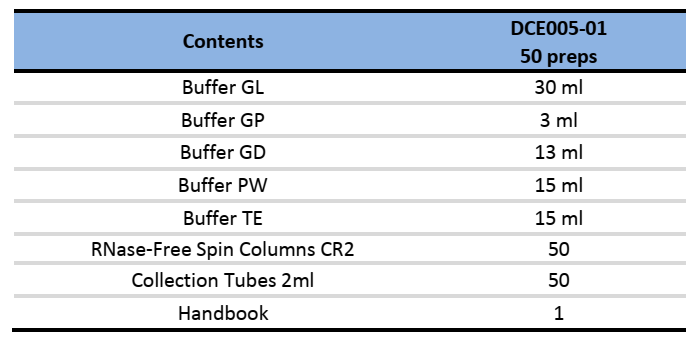
Specifications:
| Sample amount | 5-8 pieces, 5-10 μm |
| Features | Fast and convenient workflow enables DNA isolation within 1h. The unique deparaffinization solution is more safety than xylene. |
| Application | The purified DNA can be used directly for molecular biological experiments such as PCR, restriction enzyme digestion and Southern hybridization. |
| Sample type | FFPE tissues |
Ensure that ethanol (96-100%) has been added into Buffer GD and Buffer PW as indicated on the tag of the bottle before use.
- Sample preparation:
a. Paraffin section: take 5-8 paraffin sections (each section with thickness of 5 - 10 μm and surface area of 1 × 1 cm2).
b. Paraffin Block: remove excess paraffin off the sample by a scalpel, cut up the tissue sample (30 mg) into as small pieces as possible.
Note: If the sample surface has been exposed to air, discard the first 2-3 sections.
c. Samples of formalin and other fixative solutions: cut up the tissue sample (30 mg) into as small pieces as possible by a scalpel, and transfer into a 1.5 ml centrifuge tube, add 500μl PBS (10mM, pH7.4), centrifuge for 1min at 12,000 rpm (~13,400 ×g), and discard the flow-through, repeat 3 times. - Immediately place the sample in a 1.5 ml centrifuge tube. Add 500μl Buffer GL, 50μl Buffer GP, vortex vigorously for 10 s.
- Incubated at 98 °C for 30 min, mix the tube for several times during incubation, until the sample is completely dissolved.
- Centrifuge for 5 min at 12,000 rpm (~13,400 × g).
- Transfer the clear aqueous phase of the mesosphere into a new centrifuge tube.
- (optional) Add 2μl RNase A(100mg/ml), incubated for 2 min at room temperature (15-25°C).
- Add 2 volume of Ethanol (96%-100%) (For example, add 900 μl Ethanol (96%-100%) to 450 μl aqueous phase) to mix, stand for 3 min at room temperature (15-25°C).
- Transfer the mixture to Spin Columns CR2 (put CR2 into collection tube), centrifuge for 2min at 8,000 rpm (~6,000 ×g). Discard the flow-through in the collection tube, and put the spin column back to the collection tube.
- Add 500μl Buffer GD (Ensure that ethanol (96-100%) has been added) to Spin Columns CR2, centrifuge for 1min at 8,000 rpm (~6,000 ×g). Discard the flow-through in the collection tube, and put the spin column back to the collection tube.
- Add 600μl Buffer PW (Ensure that ethanol (96-100%) has been added) to Spin Columns CR2, centrifuge for 1min at 8,000 rpm (~6,000 ×g). Discard the flow-through in the collection tube, and put the spin column back to the collection tube.
- Repeat step 10.
- Put the spin column back to the collection tube, centrifuge for 2 min at 12,000 rpm (~13,400 × g), discard the flow-through in the collection tube. Open the lid and place the Spin Column CB2 at room temperature (15-25°C) for several minutes to dry membrane completely.
- Discard the Collection Tube and transfer the Spin Column CB2 to a clean 1.5 ml centrifuge tube. Pipet 30-100 μl 65°C preheated Buffer TE or ddH2O directly onto the Spin Column CB2 membrane, incubate for 2-5 min at room temperature (15-25°C), and then centrifuge for 2 min at 12,000 rpm
(~13,400 × g) to elute.



