Based on the unique silica membrane innovation and its buffer solution, the Bacteria Genomic DNA Kit is specially made for genomic DNA isolation from a wide range of Gram-negative and Gram-positive bacteria. Proteins and other organic compounds are removed to the maximum extent possible. In addition, the kit can be employed to purify genomic DNA from bacteria in food specimens. Based on the unique silica membrane innovation and its buffer solution, the Bacteria Genomic DNA Kit is particularly made for genomic DNA isolation from a wide range of Gram-negative and Gram-positive bacteria. Proteins and other organic compounds are removed to the maximum extent possible. In addition, the kit can be employed to purify genomic DNA from bacteria in food specimens.
Yield
| Sample | Quantity | DNA yield |
| Bacterial culture | 106-108cells | 5-20μg |
Storage:
Store at room temperature (15-25℃)
Components:
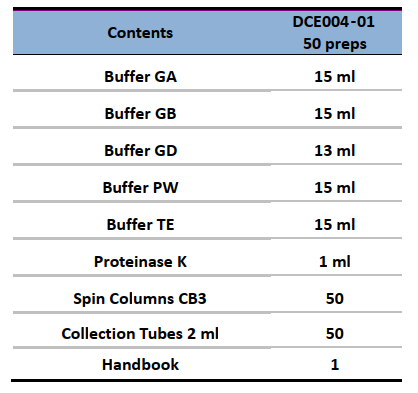
Specifications:
| Sample amount | 1-5 ml bacterial culture |
| Features | Fast and convenient workflow enables DNA isolation from Gram-negative bacteria within 1h. |
| Application | The high pure genomic DNA can be used directly in downstream experiments such as PCR, restriction enzyme digestion, Southern hybridization and DNA library construction. |
| Species Category | Gram-negative, Gram-positive bacteria, pathogenic bacteria in food |
Ensure that Buffer GD and Buffer PW have been prepared with appropriate volume of ethanol (96-100%) as indicated on the bottle and shake thoroughly.
- Pipet 1-5 ml bacterial culture suspension in a centrifuge tube, centrifuging for 1 min at 10,000 rpm (~11,500 × g), discard supernatant as possible.
- Add 200 μl Buffer GA. Mix thoroughly by vortex.
Note: For difficult-broken Gram-positive bacteria, Step 2 could be replaced by lysozyme treatment: add 110 μl enzymatic lysis buffer (20 mM Tris·Cl, pH 8.0; 2 mM sodium EDTA; 1.2% Triton® X-100), and 70 μl lysozyme (final concentration of 20 mg/ ml). Incubate for at least 30 min at 37°C. Lysozyme should be prepared with buffer, otherwise the lysozyme is not active. - Add 20 μl Proteinase K. Mix thoroughly by vortex.
- Add 220 μl Buffer GB to the sample, vortex for 15 s, and incubate at 70°C for 10 min to yield a homogeneous solution. Briefly centrifuge the 1.5 ml centrifuge tube to remove drops from the inside of the lid.
- Add 220 μl ethanol (96-100%) to the sample, and mix thoroughly by vortex for 15 s. A white precipitate may form on addition of ethanol. Briefly centrifuge the 1.5 ml centrifuge tube to remove drops from the inside of the lid.
- Pipet the mixture from step 5 into the Spin Column CB3 (in a 2 ml collection tube) and centrifuge at 12,000 rpm (~13,400 × g) for 30 s. Discard flow-through and place the spin column into the collection tube.
- Add 500 μl Buffer GD (Ensure ethanol (96-100%) has been added) to Spin Column CB3, and centrifuge at 12,000 rpm (~13,400 × g) for 30 s, then discard the flow-through and place the spin column into the collection tube.
- Add 600 μl Buffer PW (Ensure ethanol (96-100%) has been added) to Spin Column CB3, and centrifuge at 12,000 rpm (~13,400 × g) for 30 s. Discard the flow-through and place the spin column into the collection tube.
- Repeat Step 8.
- Centrifuge at 12,000 rpm (~13,400 × g) for 2 min to dry the membrane completely.
Note: The residual ethanol of buffer PW may affect downstream application. - Place the Spin Column CB3 in a new clean 1.5 ml centrifuge



