We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy


We are dedicated to providing outstanding customer service and being reachable at all times.
SSRs/STRs Analysis
Our SSRs/STRs Analysis service offers an unparalleled combination of long-read sequencing technology and sophisticated bioinformatics analysis, empowering you to unravel the mysteries hidden within repetitive genomic regions.
Overview of SSRs/STRs
Microsatellite markers, also referred to as short tandem repeats (STRs) or simple sequence repeats (SSRs), are repetitive sequences found throughout the genomes of eukaryotic organisms. These sequences consist of tandem repeat fragments (core sequences) comprised of 2-6 nucleotides in a series. SSRs contain two to six nucleotide tandem repeats (core sequences), and the number of repetitions of these repeating units varies significantly among individuals.
Repetitive sequences have been associated with over 40 neuromuscular and neurodegenerative disorders, including well-known conditions like Fragile X Chromosome, Huntington's Disease, spinal cerebellar ataxia, and others. Furthermore, microsatellite instability (MSI) is a characteristic observed in many cancer genomes. Diseases resulting from the expansion of repetitive sequences are known as repetitive sequence expansion diseases. However, it's worth noting that the shortening of repetitive sequences can also lead to certain diseases. The pathogenesis of these disorders is closely linked to the number of repeats present in these microsatellite sequences.
To detect the number of satellite sequence repeats, long read-length data can be effectively utilized. These long read lengths offer the advantage of easily spanning low-complexity regions of the genome and not displaying any GC preference, making them more suitable for detecting STRs.
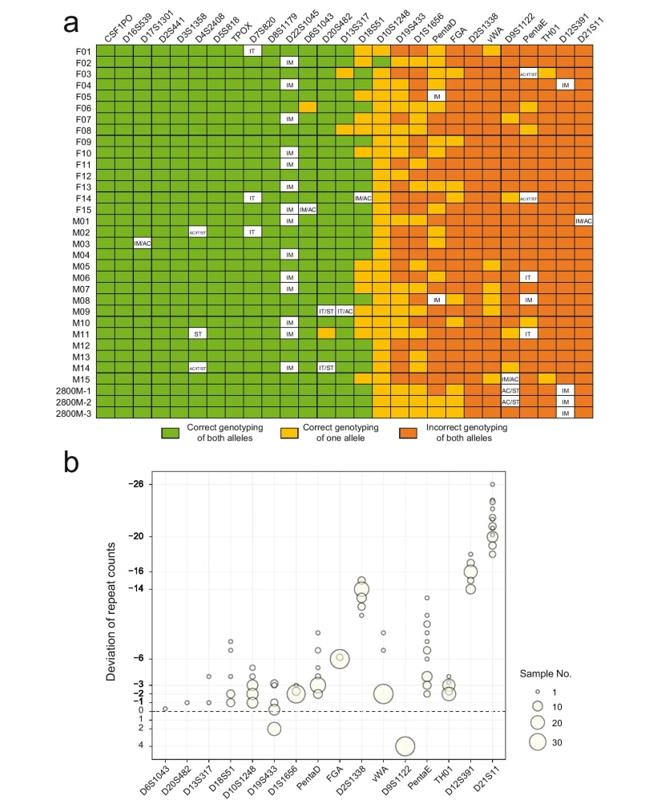 Genotyping of autosomal STRs based on MinION sequencing. (Ren et al., 2021)
Genotyping of autosomal STRs based on MinION sequencing. (Ren et al., 2021)
Features
- Precise Detection: Long-read sequencing for STR analysis represents a significant advancement, enabling precise detection of DNA sequences and revealing true disparities between core and flanking sequences of STR alleles. Unlike conventional PCR-CE methods, we can unveil distinct genotypes even in loci previously presumed to be of the same genotype. Furthermore, long-read sequencing achieves comparable discriminatory power with fewer motifs, showcasing its superior efficiency.
- Unparalleled Sensitivity: Long-read sequencing demonstrates unparalleled sensitivity in detecting DNA sequences from trace samples in hybrid assays. Its high-throughput capabilities empower the processing and analysis of vast amounts of data, enabling the identification of DNA sequences even from minute sample quantities.
- Comprehensive Coverage: Long-read sequencing offers an exceptional advantage by generating a wealth of genetic marker information from a single micro-sample experiment. This information encompasses a diverse range of genetic variations, including SNP, Indel, and mRNA data, which is unattainable with traditional sequencing platforms. Such comprehensive data mining from minuscule biological samples enhances our ability to unravel the genetics-related intricacies of a case.
Workflow of Our Short Tandem Repeats Analysis Service
 Genotyping of autosomal STRs based on MinION sequencing. (Ren et al., 2021)
Genotyping of autosomal STRs based on MinION sequencing. (Ren et al., 2021)
- Sample Preparation: We provide detailed guidelines for sample collection and preparation to ensure optimal sequencing outcomes.
- Long-read Sequencing: The prepared samples undergo cutting-edge long-read sequencing, generating data with high resolution and minimal biases.
- Data Analysis: Our experienced bioinformatics team processes the raw sequencing data, employing specialized tools for SSRs/STRs identification and variant calling.
Sample Requirements
- Sample type: Blood, tissues, cells, DNA samples and more.
- DNA amount ≥ 5 μg, Concentration ≥ 20 ng/µL, OD260/280 =1.8 ~2.0.
Our Bioinformatics Support
- SSRs/STRs Profiling: Comprehensive profiling of repetitive regions, including identification, characterization, and annotation of SSRs and STRs.
- Genotyping: Accurate determination of genotypes at targeted loci, aiding in various genetic studies and applications.
- Structural Variation Analysis: In-depth investigation of complex structural variations and repeat expansions within the genome.
- Data Interpretation: Expert analysis and interpretation of results, providing valuable insights into the implications of identified variations.
Case Studies
Detection of STR expansions Causing Neuromuscular Diseases
It is now known that over 50 of neuromuscular diseases are caused by short tandem repeat (STR) expansions, involving 37 different genes. The conventional molecular tests and short-read sequencing techniques have encountered challenges in accurately diagnosing these complex STR expansions. To address this issue, researchers present a groundbreaking approach that employs programmable targeted long-read sequencing, to simultaneously genotype all known neuropathogenic STRs in a single assay. This observation suggests that long-read sequencing has the potential to revolutionize the genetic landscape of repeat disorders, providing crucial insights into the underlying mechanisms of these diseases.
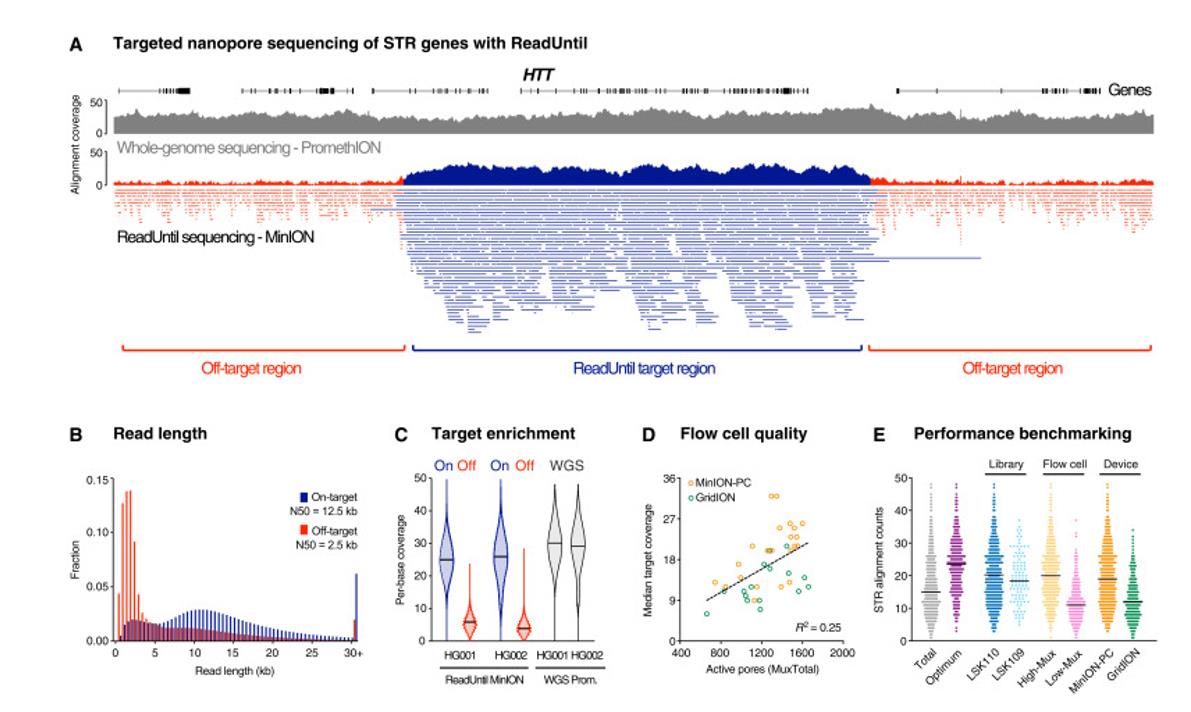 Pathogenic STR sites with ONT ReadUntil. (Ren et al., 2021)
Pathogenic STR sites with ONT ReadUntil. (Ren et al., 2021)
Reference
-
Ren, Zi-Lin, et al. "Forensic nanopore sequencing of STRs and SNPs using Verogen's ForenSeq DNA signature prep kit and MinION." International journal of legal medicine 135.5 (2021): 1685-1693.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment