We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy


We are dedicated to providing outstanding customer service and being reachable at all times.
Long Read Sequencing for Splice Variation
Splice variants play an important role in increasing proteomic diversity and cellular function within cells. Splice variants are also associated with disease states and may play a role in their etiology. To date, most information on splice variants has come from primary transcripts or through cellular studies. However, conventional short-read RNA-Seq methods usually fail to span full-length isoforms and require their computational reassembly, which may lead to erroneous reconstructions. The development of long-read sequencing technologies has provided powerful tools for studying selective splicing, heterodimer expression, genome assembly, and the detection of complex structural variants. CD Genomics is a leading global life sciences company, and we continue to expand our solutions and improve our available resources to help scientists accelerate their basic research. Based on our advanced PacBio SMRT sequencing and ONT Nanopore sequencing, and years of sequencing experience, we are committed to providing the best solutions for selective splicing variation regulation to meet the project needs of our global customers.
What Is Alternative Splicing?
Alternative splicing is an important mechanism of gene regulation. It allows a single gene to produce multiple different mRNAs, thereby greatly increasing transcriptome and proteome diversity. According to recent genome-wide association studies (GWAS), nearly 95% of protein-coding genes in the human genome undergo varying degrees of alternative splicing. Alternative splicing is highly controlled, including splice site selection, spliceosome assembly, and control of multiple splicing regulatory elements. Any disruption or mutation in the splicing machinery may affect the production of mature mRNA and functional proteins and induce various disease states. Abnormal alternative splicing can lead to various neurological diseases and cancers, and contribute to aging, infection, inflammation, immunity, and metabolic disorders. Alternative splicing may underlie the identification of novel diagnostic and prognostic biomarkers as well as new therapeutic strategies. Therefore, a deeper understanding of the regulation of alternative splicing has the potential not only to elucidate basic biological principles but also to provide solutions to various diseases.
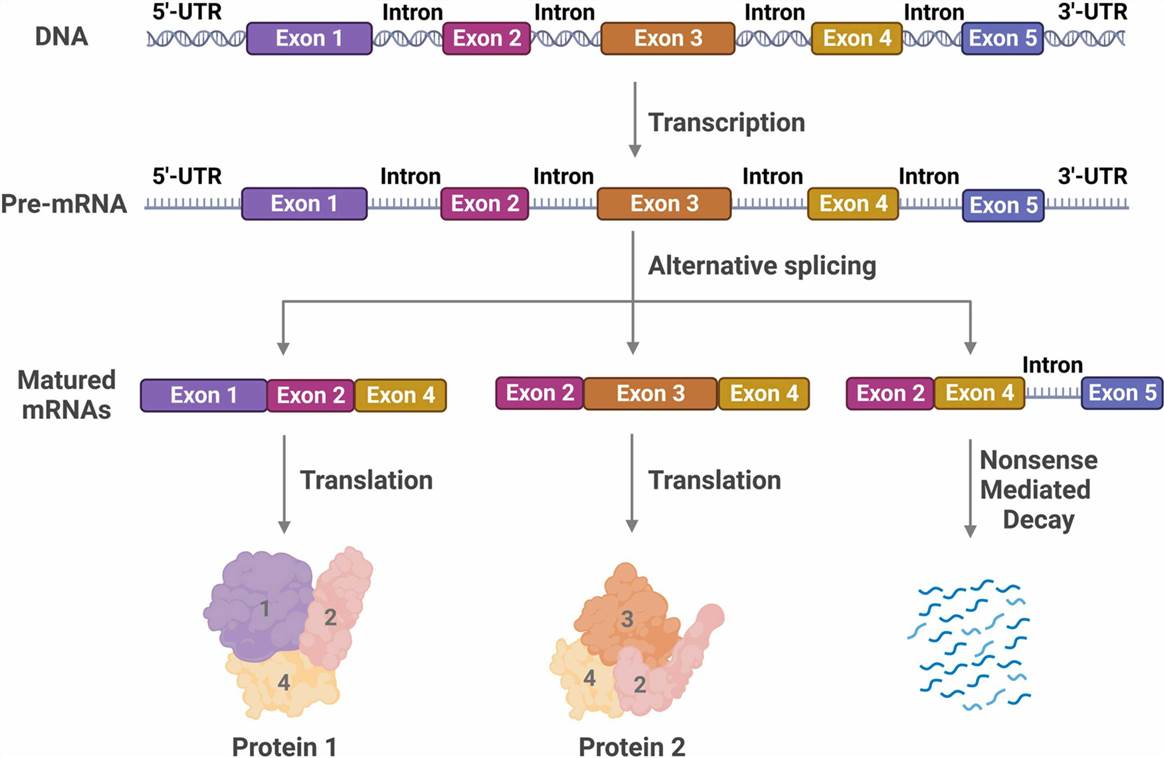 Fig. 1. Schema of alternative splicing. (Wu et al., 2021)
Fig. 1. Schema of alternative splicing. (Wu et al., 2021)
Solutions Offering at CD Genomics
Traditional short-read sequencing, although widely used, has limitations in comprehensive splice variant analysis, in particular its inability to efficiently reconstruct full-length isoforms. CD Genomics provides professional long read sequencing solutions for splice variants, allowing full-length transcript recovery and quantification, enabling transcript-level analysis of alternative splicing processes and how these processes change with cellular states.
We utilize advanced long-read sequencing to ensure that the read length is exactly equal to the fragment length. This method allows the sequencing of transcripts larger than 20 kb in a single read. With such capabilities, identification and quantification of entire isomers become more streamlined and precise.
In addition to standard cDNA molecules, our technology can directly sequence native RNA molecules. This dual role not only retrieves nucleotide sequence data but also obtains base modification information.
- Study Protein Isoform Diversity
Long-read sequencing also enables the study of protein isoform diversity, which is primarily the result of alternative splicing at the RNA level. We develop long-read proteomics pipelines for integrating sample-matched long-read RNA sequencing and mass spectrometry-based proteomic data to enhance the detection and classification of protein isoforms. In this analysis process, long-read RNA sequencing data is used to predict full-length protein isoforms and generate a database, which can be used as a reference for mass spectrometry data analysis.
Competitive Advantages
- Not only can exon junctions be observed, but entire isoforms can be resolved with unparalleled clarity, making it possible to obtain higher-resolution isoforms.
- Identifying epigenetic modifications of nucleotide sequences by direct RNA sequencing.
- Streamlined workflows ensure the highest fidelity of data generated.
- Accurately resolve and quantify full-length splice variants from long sequencing reads.
- Meet your output requirements with a range of sequencing platforms.
- Helping to reveal previously unknown areas, particularly in genes associated with neuropsychiatric diseases.
With its unparalleled expertise and cutting-edge technology, CD Genomics is a trusted partner to the genomics community, driving the advancement of alternative splicing research. Our goal is to improve the clinical interpretation of non-canonical splice variants to increase the diagnostic yield of whole-genome sequencing data. If you have any questions, please feel free to contact us. We look forward to working with you on projects of interest.
References
- Wu, Siyuan, and Ulf Schmitz. "Single-cell and long-read sequencing to enhance modelling of splicing and cell-fate determination." Computational and Structural Biotechnology Journal (2023).
- Clark, Michael B., et al. "Long-read sequencing reveals the complex splicing profile of the psychiatric risk gene CACNA1C in human brain." Molecular psychiatry 25.1 (2020): 37-47.
Related Services
PacBio SMRT Sequencing
Oxford Nanopore Sequencing
Full-Length Transcript Sequencing (Iso-Seq)
Single-Cell Full-Length Transcriptome Sequencing
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment