We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy


We are dedicated to providing outstanding customer service and being reachable at all times.
PacBio HiFi Sequencing for Spinal Muscular Atrophy Analysis
At a glance:
- Overview
- Paraphase Identifies Full-Length SMN1 and SMN2 Haplotypes
- Benefits of PacBio HiFi Sequencing for SMA Analysis
Overview
Spinal muscular atrophy (SMA) has long been recognized as a serious neuromuscular disease, is currently ranked as the leading cause of early infant mortality, and is one of the most common recessive diseases worldwide. Two genes play a role in the onset and severity of SMA: SMN1 and SMN2. SMN1 is highly homologous to its paralog SMN2. These two genes are located in genomic regions with highly complex long repetitive sequences that are nearly identical except for a few bases, which makes sequence analysis and variant calling challenging. Both genes have different copy numbers in different populations and have been analyzed by a variety of methods (usually PCR-based dosage testing combined with sequencing), each of which has its limitations. Further, without pedigree information, it is currently not possible to identify silent carriers (2+0) with two copies of SMN1 on one chromosome and zero copies on the other.
To better facilitate SMA screening, there is an urgent need for a method that allows for comprehensive whole-gene SMN1 and SMN2 analysis. Ideally, the method should be able to (1) identify copy numbers of intact SMN1 and SMN2 based on c.840, (2) identify pathogenic variants in SMN1 other than c.840C loss, and (3) identify silent carriers. Accurate long-read PacBio HiFi sequencing is well suited for resolving regions of high sequence homology and has been demonstrated in an amplicon-based study of a Chinese population. A cutting-edge tool, Paraphase, accurately detects copy number and variation across SMN1 and SMN2 using PacBio HiFi sequencing.
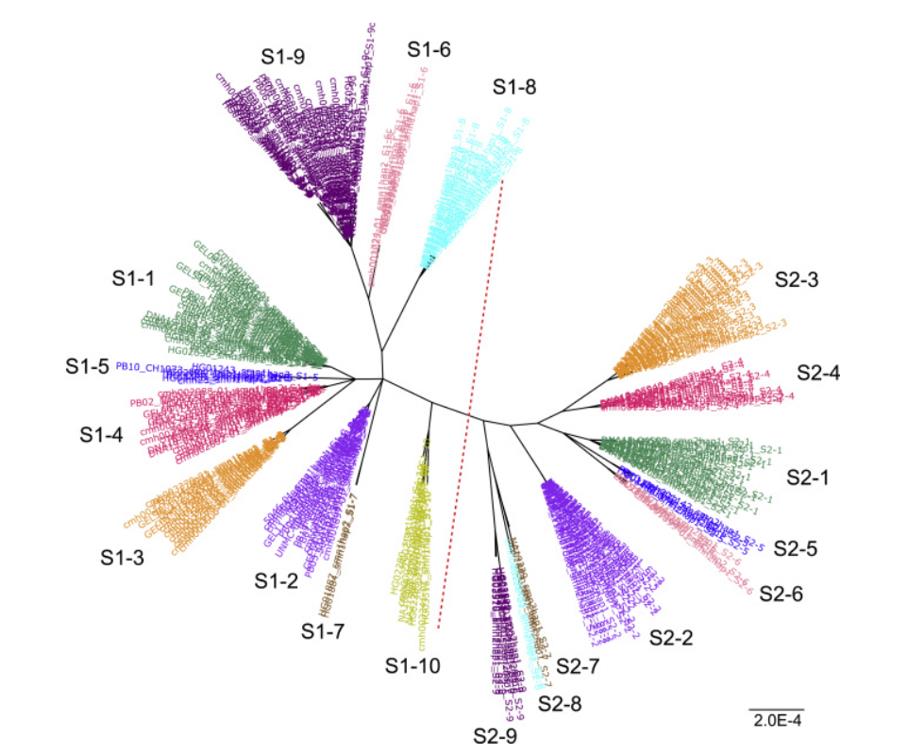 Population-wide haplotype analysis identified major SMN1 and SMN2 haplogroups. (Chen et al., 2023)
Population-wide haplotype analysis identified major SMN1 and SMN2 haplogroups. (Chen et al., 2023)
Paraphase Identifies Full-Length SMN1 and SMN2 Haplotypes
Advances in sequencing technology have given us the power of PacBio HiFi sequencing. This approach has revealed unprecedented insights into the SMA genetic landscape, and the innovative software tool Paraphase has been an integral part of this revolution.
Paraphase is an informatics approach that identifies full-length SMN1 and SMN2 haplotypes, determines gene copy numbers, and calls fixed-phase variants using long-read PacBio HiFi data. For example, one study applied Paraphase to 438 samples from five ethnic populations and performed a population-wide haplotype analysis of these highly homologous genes. This study identified the major SMN1 and SMN2 haplogroups and characterized their co-segregation through pedigree-based analysis. Beyond simple copy number testing, Paraphase can detect pathogenic variants and enable potential haplotype-based silent carrier screening by statistically phasing haplotypes into alleles.
Benefits of PacBio HiFi Sequencing for SMA Analysis
- Accuracy and consistency: PacBio HiFi sequencing stands out for its high consistency and reliability, especially when cross-referenced with other established methods. Paraphase's gene copy number analysis has been shown to match alternative methods with up to 99.2% accuracy for SMN1 and 100% accuracy for SMN2.
- Comprehensive Analysis: PacBio HiFi sequencing, when used with Paraphase, provides a broader understanding of SMA than just copy numbers. It can detect disease-causing variants throughout the gene, providing insight into the more subtle genetic complexities of the disease.
- Potential for vector screening: Perhaps one of the most promising advantages is the tool's potential to enhance vector screening. The discovery of specific haplotypes that form double-copy SMN1 alleles could greatly improve the accuracy of silencing vector assays landmark advance in SMA research.
Reference
- Chen, Xiao, et al. "Comprehensive SMN1 and SMN2 profiling for spinal muscular atrophy analysis using long-read PacBio HiFi sequencing." The American Journal of Human Genetics. 110.2 (2023): 240-250.
Related Services
PacBio SMRT Sequencing Technology
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment