We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy


We are dedicated to providing outstanding customer service and being reachable at all times.
What is the RNA Integrity Number (RIN)?
At a glance:
- Overview
- What is RNA Integrity Number
- The Rationale for RIN Assays
- What is An Acceptable RIN Score
- How is RIN Calculated
- The Importance of RIN in Molecular Biology
- What RIN Can Do
- Limitations of RIN
- Factors Affecting RIN
Overview
At CD Genomics, we pride ourselves on the quality of the long read long sequencing data we generate for Pacific BioSciences (PacBio) and Oxford Nanopore Technology (ONT). High-quality sequencing data is the ultimate goal of next-generation sequencing. Since the final quality is largely dependent on the quality of the starting material, it is important to ensure high-quality starting material.
Unlike DNA, RNA is a highly sensitive nucleic acid that can be easily degraded if not handled properly. Other factors such as heat, contaminated chemicals, and inadequate buffer conditions can also affect the integrity of RNA and lead to degradation. Evaluating its quality and integrity is therefore a critical step in ensuring its suitability for downstream processes. the gold standard for RNA integrity analysis is the RNA Integrity Number (RIN), which indicates the degree of integrity of the RNA. A RIN score of >8.0 is ideal for high throughput downstream processing.
What is RNA Integrity Number
RIN is a numerical value that indicates RNA integrity. Technically, it applies to microfluidic chip-based isolation and allows us to visualize bands and measure integrity values by calculation and using mathematical models.
There are several methods available for measuring RNA quality, such as 260/280 ratios, electrophoretic analysis, and banding patterns of 28S and 18S rRNA fragments, but the results are not guaranteed to be very accurate. The technique is based on the principle of conventional gel electrophoresis using EtBr stained RNA. By observing the banding pattern as the RNA migrates through the gel, RNA integrity can be more fully assessed.
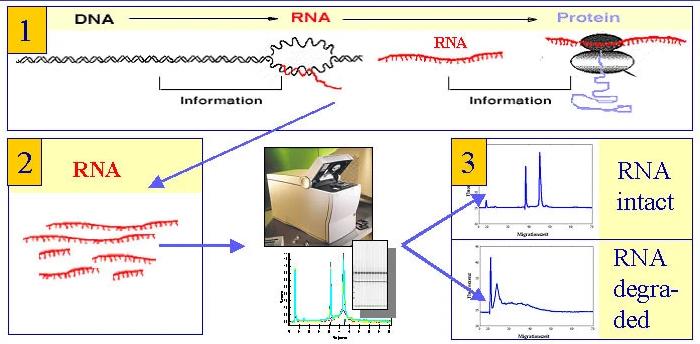 Application environment. (Schroeder A et al., 2006)
Application environment. (Schroeder A et al., 2006)
The Rationale for RIN Assays
The principle behind the RIN assay is rooted in a method developed by Agilent Technologies that utilizes the capabilities of capillary electrophoresis (CE) and Bayesian adaptive learning methods. In this method, smaller volumes of RNA samples are run in smaller-sized capillaries and separated by capillary electrophoresis.
The resulting electropherogram displays the RNA fragment bands as peaks that emit fluorescence with an intensity proportional to the peak size for the particular RNA species. Thus, the larger the RNA fragments, the more fluorescence is emitted and the higher the intensity of the bands. This principle makes it possible to clearly and accurately distinguish between different-sized nucleic acid fragments, which helps in the accurate calculation of RIN.
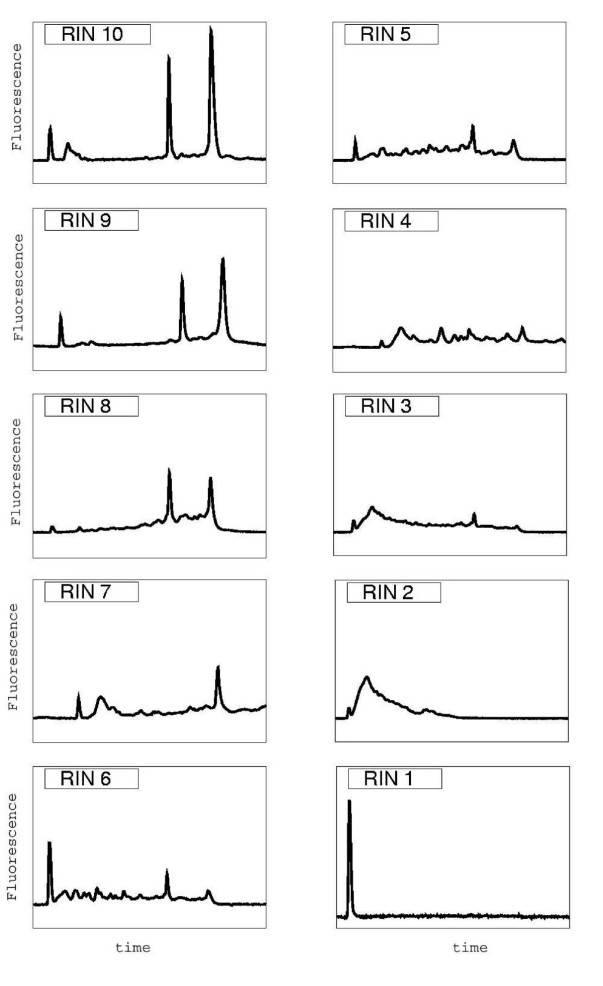 RNA integrity categories. (Schroeder A et al., 2006)
RNA integrity categories. (Schroeder A et al., 2006)
What is an Acceptable RIN Score
In the RIN system, RNA integrity is expressed on a scale of 1 to 10, where 10 indicates excellent RNA integrity and 1 indicates severe RNA degradation. However, understanding what constitutes an "acceptable" RIN score is critical for proper application in molecular biology research.
In general, RIN scores between 8 and 10 are considered to indicate highly intact RNA, making them ideal for most research applications. On the other hand, scores between 1 and 5 indicate low RNA integrity or a high degree of degradation, generally making them unsuitable for many experiments. Moderately intact or lowly degraded RNA is usually indicated by a RIN score between 6 and 8. Therefore, in most cases, a RIN score of 7 to 10 is considered acceptable.
However, it is worth noting that different molecular genetic tests have different RIN score requirements:
- Gene arrays: RIN score 6 to 8
- RT-qPCR: RIN score 5 to 6
- qPCR: RIN score >7
- RNA Sequencing: RIN score between 8 and 10
- Microarray: RIN score between 7 and 10
How is RIN Calculated
The RIN is calculated using a multifaceted approach that takes into account several parameters. The foundation of the process is based on capillary electrophoresis, which allows smaller volumes of RNA samples to be run in smaller-sized capillaries and separated accordingly. The separation produces an electropherogram that represents the RNA fragment bands as peaks.
To better understand the RNA sample, the RIN algorithm includes not only the 28S, 18S, and 5S rRNA peaks, but also any anomalies in the labeled and fast regions. Essentially, it provides exhaustive electrophoretic profiling beyond the major rRNA bands. This level of comprehensive analysis helps to accurately and objectively calculate RIN values without the need for manual calculations or equations, thus ensuring consistent measurements.
The Importance of RIN in Molecular Biology
RIN plays a vital role in molecular biology. It acts as a gatekeeper, determining the suitability of RNA samples for further processing, which is essential for gene expression, RNA sequencing, and microarray studies. High-quality RNA is critical to the success of such experiments, and the RIN is a key player in confirming the suitability of RNA samples.
RIN can help optimize the use of resources and improve the reliability and reproducibility of results by efficiently assessing the quality of RNA before starting expensive and labor-intensive experiments. It is an important tool for preventing experimental failure due to RNA sample degradation, saving time, effort, and money.
What RIN Can Do
- Get an assessment of RNA integrity.
- Directly compare RNA samples (e.g., before and after shipment, compare the integrity of the same tissue in different laboratories, etc.).
- Ensure reproducibility of experiments (e.g. if the RIN shows a given value and is suitable for microarray experiments, then the same value of RIN can always be used for microarray experiments if the same organism/tissue/extraction method is used).
Limitations of RIN
Although RIN provides a great deal of information about RNA integrity, it is not without limitations. For example, without prior validation, RINs cannot predict the success of an experiment. This means that an RNA sample with a particular RIN may be suitable for one type of experiment but not for another.
Factors Affecting RIN
RNA Extraction Protocols
Protocols that effectively inactivate RNase tend to produce higher RIN scores. The type of tissue used also affects the RIN. Some tissues are enriched in RNase or difficult to process, which may negatively affect the RIN score.
RNA Elution Process
RNA needs to be stabilized after elution and stored properly to maintain its integrity. Failure to do so may result in decreased RNase activity, which may degrade the RNA, thereby decreasing the RIN score.
Concentration of RNA Extract
According to Agilent, RNA concentrations greater than 50 ng/μL typically produce uniform RIN scores. However, due to potential inconsistencies, it is not recommended that concentrations below 25 ng/μL be used to score the RIN.
Reference
- Schroeder, Andreas, et al. "The RIN: an RNA integrity number for assigning integrity values to RNA measurements." BMC molecular biology 7.1 (2006): 1-14.
Related Services
Oxford Nanopore Sequencing
PacBio SMRT Sequencing
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment

