We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy


We are dedicated to providing outstanding customer service and being reachable at all times.
PacBio SMRT Sequencing for Methicillin-Resistant Staphylococcus aureus (MRSA)
At a glance:
- Overview
- Mechanisms and Pathophysiology of MRSA
- Diagnosis, Screening, and Prevention of MRSA
- PacBio SMRT Sequencing for MRSA
Overview
Staphylococcus aureus is a Gram-positive, non-motile, coagulase-positive globular bacterium of the phylum Thicket. Although the genus Staphylococcus includes 52 species and 28 subspecies, S. aureus is by far the most clinically relevant. Staphylococcus aureus is present in 20-40% of the nasal mucosal commensal microbiota of the general population. When the skin and mucosal barriers are disrupted, such as due to chronic skin disease, wounds, or surgical interventions, Staphylococcus aureus can enter the underlying tissues or bloodstream and cause infection. Individuals who use invasive medical devices (e.g., peripheral and central venous catheters) or have compromised immune systems are particularly susceptible to Staphylococcus aureus infections. Since the 1960s, methicillin-resistant Staphylococcus aureus (MRSA) has emerged and spread globally and is the leading cause of hospital-associated (HA-MRSA) and community-associated (CA-MRSA) infections. Some of these clones have become major causes of hospital-acquired infections, blurring the distinction between CA-MRSA and HA-MRSA. Although most studies to date have originated in industrialized countries such as the United Kingdom and the United States, there is increasing awareness of the emergence of CA-MRSA clones in non-industrialized and newly industrialized countries such as Africa, Asia, and India.
MRSA is an opportunistic pathogen that can cause a range of infections, from minor skin infections to serious diseases such as pneumonia, endocarditis, and sepsis. With its growing resistance to conventional antibiotics, there is an urgent need for advanced tools and methods to study its genome, epidemiology, and virulence mechanisms. Although treatment options for MRSA are limited, several new antimicrobial agents are in development. Understanding the dynamics of colonization, routes of transmission, risk factors for infection progression, and conditions that promote the emergence of resistance will help optimize strategies for effective MRSA control. Vaccine candidates are also in development and may be an effective preventive measure. PacBio SMRT sequencing is a cutting-edge genomic tool that provides unprecedented clarity for sequencing bacterial genomes, including MRSA.
Mechanisms and Pathophysiology of MRSA
MRSA Colonization
The primary site of MRSA colonization is the nasal cavity. Longitudinal studies have demonstrated three distinct patterns, the understanding of which is critical to assessing the risk and potential spread of MRSA infection.
- Persistent carriers: This group comprises approximately 15% of individuals and carries MRSA continuously.
- Intermittent carriers: Comprised of 70% of individuals, this group experiences sporadic MRSA colonization and can spontaneously clear it.
- Non-carriers: This 15% segment never exhibits MRSA colonization.
Factors Influencing Colonization
Host genetics and the nasal microbiota both influence MRSA colonization. Polymorphisms in inflammation-related genes can significantly determine an individual's carrier status. In addition, the nasal microbiota, especially species such as Corynebacterium spp. and Staphylococcus epidermidis, play a key role in MRSA colonization, often competing for nutrient and adhesion sites.
Regulation of MRSA Virulence
Once inside the body, MRSA adheres to host epithelial cells. In the initial phase, phosphomimetic acids on the bacterial cell wall facilitate this adhesion, followed by microbial surface components that recognize molecules of the adhesion matrix. Among these, Staphylococcus aureus aggregation factor B (ClfB) plays a crucial role in promoting adhesion and subsequent colonization.
After colonization, MRSA is not idle. It actively participates in and in many cases circumvents the host's immune response. For example, although abscess formation is the primary host defense mechanism, MRSA can adopt strategies to survive within the abscess. It does this by producing toxins that prevent leukocytes (especially neutrophils) from destroying the bacteria. This action allows methicillin-resistant Staphylococcus aureus to proliferate, leading to more serious infections and complications.
MRSA is able to cause a wide range of infections because it contains a large number of virulence factors. These factors may have adhesive properties that can damage host cells and even modulate the host's immune response. Examples include surface proteins that promote adhesion to injured tissue and cytolytic toxins that damage host cells.
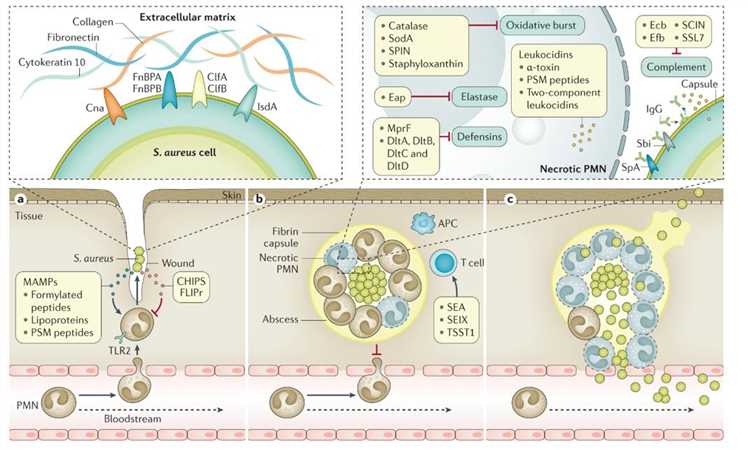 Stages of Staphylococcus aureus infection. (Lee et al., 2018)
Stages of Staphylococcus aureus infection. (Lee et al., 2018)
Mechanisms of Methicillin Resistance
At the heart of MRSA's notoriety is its resistance to methicillin and other β-lactam antibiotics. This resistance is primarily conferred by the mecA gene, which encodes penicillin-binding protein 2a (PBP2a). PBP2a has a very low affinity for most β-lactam antibiotics. When bacteria are exposed to these antibiotics, PBP2a takes over the important function of peptidoglycan biosynthesis, allowing MRSA to survive and multiply in the presence of the drug.
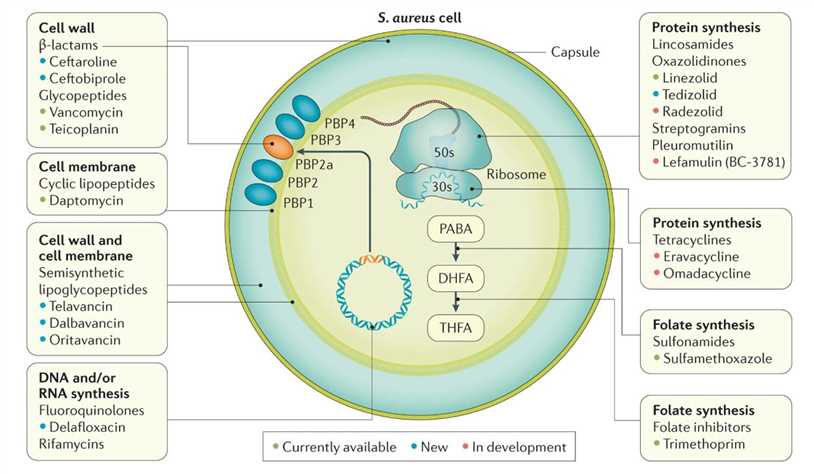 Bacterial targets of antibiotics active against MRSA. (Lee et al., 2018)
Bacterial targets of antibiotics active against MRSA. (Lee et al., 2018)
Diagnosis, Screening, and Prevention of MRSA
Diagnosis of MRSA
Phenotypic Methods
Phenotypic methods serve as the gold standard for MRSA identification. One such method is the disk-diffusion approach. Here, a cefoxitin disk is placed on Mueller-Hinton agar. If MRSA is present, it showcases resistance, evident by the clear zone (or lack thereof) around the disk. This approach is sensitive, especially with the incorporation of cefoxitin, which acts as a better inducer for MRSA-associated genes (mecA and mecC) compared to methicillin.
Non-Phenotypic Methods
A more contemporary approach involves matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF MS). This technology analyzes the bacterial protein profile, comparing it to a robust database, ensuring accurate identification. However, the accuracy of MALDI-TOF MS heavily relies on the purity and quantity of the microorganism sample. Furthermore, DNA-based techniques like multiplex real-time PCR assays have emerged as dependable tools, offering results in a relatively shorter timeframe - usually within 1.5 hours.
MRSA Screening
Chromogenic Media
Chromogenic media, with their ability to display distinct colony colors based on the enzymatic activity of the bacteria, have evolved as primary diagnostic tools. The combination of chromogenic media with advanced technologies like MALDI-TOF MS has further enhanced the diagnostic process, both in terms of specificity and speed.
PCR-Based Assays
Real-time PCR-based assays, especially when employed for nasal swab screening, can significantly reduce the waiting period for results. Such assays can detect MRSA within 1-2 hours, a marked improvement over the 14-18 hours necessitated by chromogenic media-based tests.
Prevention of MRSA Spread
Emphasizing Hand Hygiene
Among the preventive measures, hand hygiene stands out as the most critical. By ensuring proper hand hygiene, the transmission of MRSA can be significantly curtailed. The World Health Organization (WHO) underscores the importance of this simple yet impactful measure, providing exhaustive guidelines to guarantee optimum hand hygiene practices in healthcare settings.
Active Surveillance
Proactive screening or active surveillance aids in the identification of asymptomatic MRSA carriers, enabling timely interventions. Depending on the circumstances, MRSA screening can be universal (covering all patients) or targeted (restricted to high-risk groups). While the efficacy of universal screening remains debated, it's essential to align any change in screening strategy with the local MRSA epidemiology and patient vulnerability.
Contact Precautions and Isolation
The usage of contact precautions, which entails the donning of disposable gowns and gloves, is widely endorsed when catering to MRSA-positive patients. This measure considerably curtails the risk of transmission. Additionally, isolating MRSA-positive patients in single rooms is a practice adopted by numerous healthcare facilities, although its effectiveness in some scenarios, especially in intensive care units with endemic MRSA and low hand hygiene compliance, is still under discussion.
PacBio SMRT Sequencing for MRSA
MRSA is highly clonal and often escapes traditional molecular typing methods such as pulsed-field gel electrophoresis (PFGE) and Staphylococcus aureus protein A (spa) typing. While these techniques allow for rapid screening, they typically lack the resolution to distinguish closely related MRSA strains. Indeed, the clonal characterization of MRSA means that small genetic changes can lead to significant changes in virulence and antibiotic resistance. This calls for more powerful, higher-resolution technologies to understand the subtle genetic differences in MRSA strains.
Pacific Biosciences (PacBio) Single Molecule Real-Time (SMRT) Sequencing provides long-read, long-sequencing technology that significantly improves genomic analysis of MRSA. The advantage of SMRT sequencing is its ability to cover highly repetitive regions of the genome, providing accurate sequencing of challenging regions.
Comprehensive Genome Analysis
The MRSA genome consists of core and accessory elements. While the core genome remains relatively conserved, the accessory genome, which contains many of the determinants of virulence and drug resistance, can evolve rapidly. The auxiliary genome also contains repetitive sequences, such as prophages and mobile genetic elements. These highly repetitive elements pose a challenge for short read sequencing platforms. SMRT sequencing with long read lengths can span these repetitive regions, ensuring accurate sequencing of these critical regions.
Characterizing Outbreaks and Tracking Evolution
Understanding the lineage and evolutionary trajectory of MRSA strains is critical when dealing with outbreaks in hospitals. SMRT sequencing not only provides a detailed understanding of the genomic composition of MRSA strains but also helps to track microevolutionary changes, which are critical to understanding the dynamics of outbreaks.
Unraveling the Auxiliary Genome
One of the major problems with MRSA is its potential virulence. While many virulence factors are well documented, the rapid evolution of auxiliary genomes means that new virulence determinants may be emerging. With SMRT sequencing, researchers can delve deeper into the auxiliary genome to discover hidden virulence factors and gain insight into their interactions.
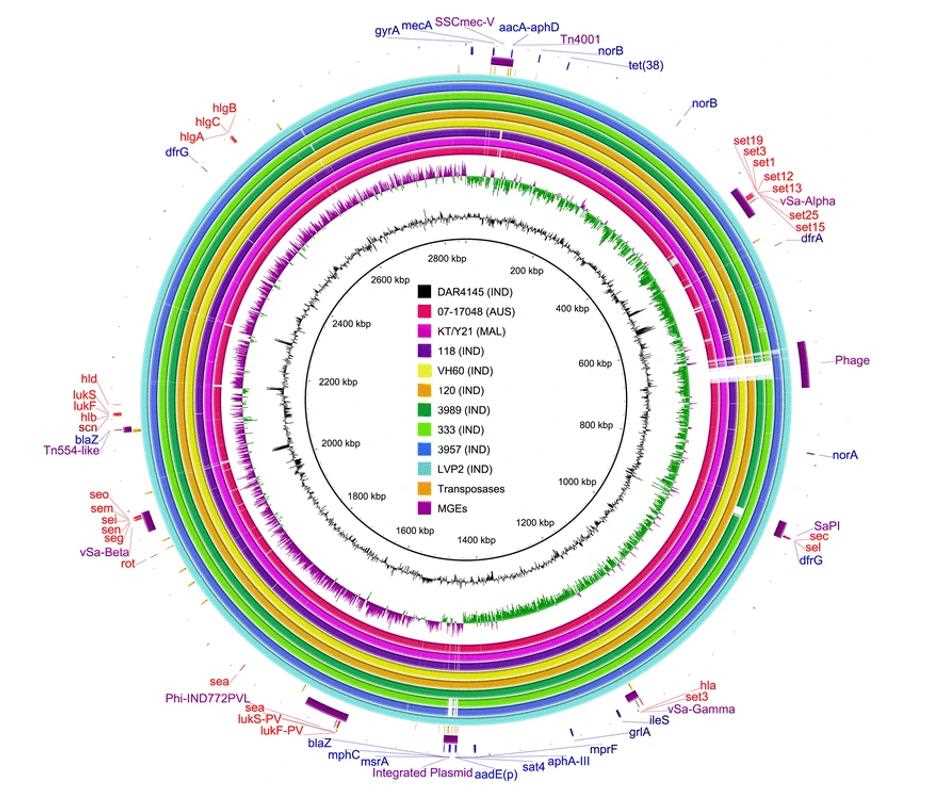 Comparison of draft genomes of nine strains of ST772-MRSA-V to the complete reference genome of strain DAR4145. (Lee et al., 2018)
Comparison of draft genomes of nine strains of ST772-MRSA-V to the complete reference genome of strain DAR4145. (Lee et al., 2018)
References
- Lee, Andie S., et al. "Methicillin-resistant Staphylococcus aureus." Nature reviews Disease primers. 4.1 (2018): 1-23.
- Steinig, Eike J., et al. "Single-molecule sequencing reveals the molecular basis of multidrug-resistance in ST772 methicillin-resistant Staphylococcus aureus." BMC genomics. 16 (2015): 1-10.
- McClure, Jo-Ann, and Kunyan Zhang. "Complete genome sequence of the methicillin-resistant Staphylococcus aureus colonizing strain M92." Genome announcements. 5.23 (2017): 10-1128.
Related Services
PacBio SMRT Sequencing Technology
Bacterial Whole-Genome Resequencing
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment