We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy


We are dedicated to providing outstanding customer service and being reachable at all times.
Long-read Sequencing Reveals Unseen Genomic Variants
At a glance:
- Overview
- Advantages of Long-read Sequencing for Genetic Variation
- Sample Requirements of Long-read Sequencing for Genetic Variation
- Applications of Long-read Sequencing for Genetic Variation
- Workflow of Long-read Sequencing for Genetic Variation
Overview
A complete map of genomic variation is important to gain insight into genetic characterization and to aid in precision disease research, which can further address research areas such as evolution, agriculture, and medicine. Although short-read long sequencing can accurately detect single nucleotide polymorphisms (SNPs), insertion or deletion changes (InDel) occurring in gene sequences for smaller types of variants, it has limited sensitivity for detection of copy number variants (CNVs) and structural variants (SVs).
In recent years, with the breakthrough innovation of sequencing platforms, long-read-length sequencing has been widely used in genome research, which is able to effectively analyze complex genome structures, including SV, CNV, and DNA-protein interaction studies, providing a more comprehensive genomic perspective for finding disease-related variants. The high accuracy of long-read-long technology also makes it possible to discover rare variants that may have been missed by short-read-long sequencing technology, providing more precise genetic variant information for basic research in precision medicine.
Long-read sequencing sequences natural DNA molecules, generating reads 10-1 million base pairs in length and providing information on DNA methylation. Oxford Nanopore Technologies (ONT) and Pacific Biosciences (PacBio) have developed long-read sequencing technologies that provide a comprehensive understanding of genomic variation. We provide state-of-the-art PacBio SMRT Sequencing and Oxford Nanopore Sequencing platforms to help you reveal unseen genomic variants.
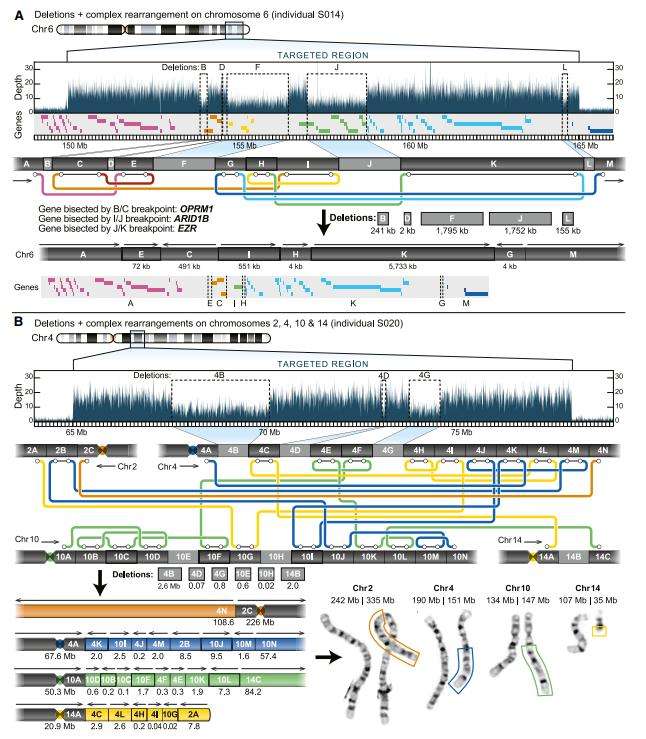 Targeted long-read sequencing identifies additional structural differences not observed by standard clinical testing. (Miller DE et al., 2021)
Targeted long-read sequencing identifies additional structural differences not observed by standard clinical testing. (Miller DE et al., 2021)
Advantages of Long-read Sequencing for Genetic Variation
- Comprehensive coverage of genetic variation: Capable of capturing a wide range of variant types, including SNPs, InDels, SVs, CNVs, and even complex methylation information.
- High accuracy and sensitivity: It can reveal rare variants that are often missed by short-read-long sequencing methods. It brings new clarity to the study of genetic variation and opens the door to a more detailed understanding of complex genetic diseases.
- Cost-effective approach: Makes it possible to obtain more comprehensive data at a lower price.
- Superior read length and uniform coverage: With its single-molecule sequencing approach, it avoids PCR amplification bias and ensures more even and consistent coverage across the genome, including regions with high GC content.
Sample Requirements of Long-read Sequencing for Genetic Variation
- Genomic DNA: The first requirement is unamplified genomic DNA. Ideally, this DNA should have a high degree of integrity, with an input volume of more than 5 µg to ensure complete coverage.
- Diversity of sample types: In addition to genomic DNA, the flexibility of long-read, long-sequencing allows for the analysis of a large number of samples, including blood, tissues and different cell lines.
Applications of Long-read Sequencing for Genetic Variation
Complex structural variation studies
Complex structural variants that are difficult to detect with short read lengths can be detected, including inversions, deletions, or translocations of large segments, some of which are associated with areas such as genetic diseases. We offer specialized human genome structural variation detection services to comprehensively detect SVs at high resolution, including large insertions, deletions, inversions, duplications, translocations, and complex combinations of these mutations.
Disease research
Can help diagnose cancers and rare diseases caused by structural variants, such as Mendelian inheritance diseases, Carney complex, etc.
Population studies
Previous population-level studies, including genome-wide association studies, have not yet fully resolved the genetic factors behind human traits and diseases. Advances in sequencing technology and bioinformatics have paved the way for population-level long-read sequencing studies.
Workflow of Long-read Sequencing for Genetic Variation
- Quality Control (QC) for Reads: The PacBio and Oxford Nanopore Technologies (ONT) platforms ensure the highest quality of raw data.
- Alignment to reference genomes: Once quality control is performed, reads are aligned to a reference genome (e.g. hg38). Tools such as Minimap2, BLASR and NGMLR play a key role in ensuring that each read finds the right place on the reference genome.
- Revealing the SV: Tools such as Sniffles, SMRT-SV, PB-Honey and NanoSV are widely used for SV detection, each with its own proprietary algorithm and detection mechanism. They ensure that various SVs are captured with minimal error.
- Organizing the data: After detection, the detected SVs must be sorted in a consistent manner. VCFsort is the tool of choice for this purpose and ensures that the detected variants are systematically organized.
- Summarizing the findings: Finally, the awk scripting language steps in to provide a concise summary of the variant calls.
References
- Miller DE, Sulovari A, Wang T, et al. Targeted long-read sequencing identifies missing disease-causing variation. Am J Hum Genet. 2021 Aug 5;108(8):1436-1449.
- De Coster W, Weissensteiner MH, Sedlazeck FJ. Towards population-scale long-read sequencing. Nat Rev Genet. 2021 Sep;22(9):572-587.
Related Services
PacBio SMRT Sequencing
Oxford Nanopore Sequencing
Human Genome Structural Variation Detection
Human Whole Genome Sequencing
Population Evolution
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment




