We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy


We are dedicated to providing outstanding customer service and being reachable at all times.
High-Molecular-Weight DNA Extraction for Long-Read Sequencing
At a glance:
- Overview
- DNA Sample Requirements
- Amount of DNA Required
- DNA Extraction Protocol
- DNA Quality Assessment
Long-read sequencing technology requires high molecular weight DNA of sufficient purity and integrity because the quality of the DNA starting material will be directly reflected in the sequencing results. Any irreversible DNA damage present in the input material (e.g., interstrand cross-links, etc.) will result in compromised performance and reduced read length. Contamination present in the sample not only affects the enzymatic reactions in library preparation but also affects the efficiency of well occupancy in sequencing, thereby reducing data throughput. High-quality, high-molecular-weight genomic DNA is critical to obtain long reads and ensure optimal sequencing performance.
Overview
Rapid advances in long-read sequencing technology have transformed read lengths from bps to Mbps, enabling chromosome-scale genome assembly. However, read length is now being limited by the extraction of pure high molecular weight DNA suitable for long-read sequencing, which is particularly challenging in plants and fungi. Extraction of high molecular weight DNA from plant material is challenging. First, the cell wall of plant cells is composed of polysaccharide polymers such as cellulose and pectin, as well as glycoproteins and lignin, making the cell wall hard and difficult to break. Therefore, steps to achieve efficient mechanical disruption of the cell wall are necessary. Plants also produce polysaccharides and phenols as chemical defenses against herbivores, which tend to accumulate in leaves and can bind DNA during cell lysis and affect downstream analyses. There are currently a variety of protocols and kits available for high molecular weight DNA extraction. In short, in order to fully realize the potential of long-read sequencing, it is necessary to obtain high molecular weight DNA of sufficient purity and integrity.
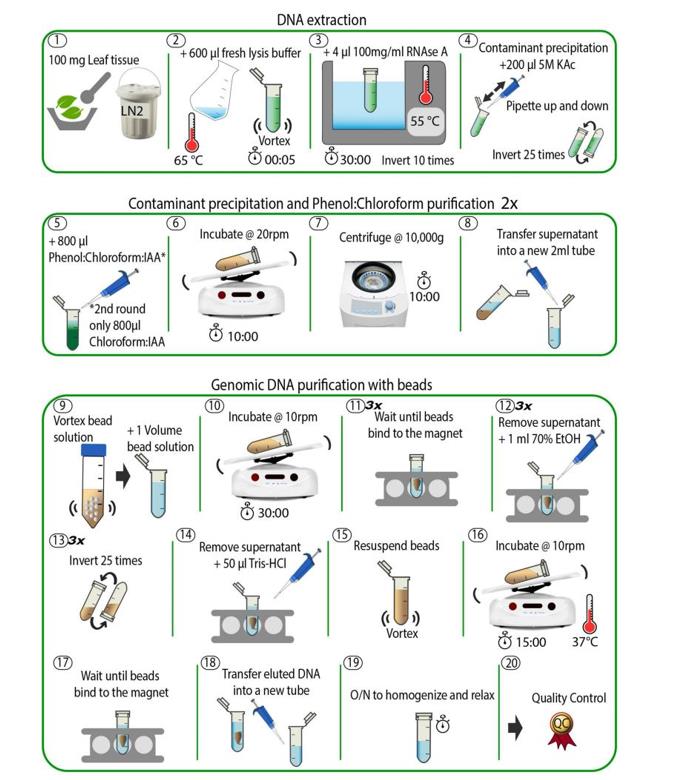 Schematic overview of the DNA extraction method. (Jebb et al., 2020)
Schematic overview of the DNA extraction method. (Jebb et al., 2020)
DNA Sample Requirements
- Is double-stranded.
- Not subjected to multiple freeze-thaw cycles as this may cause DNA damage.
- Has not been exposed to high temperatures (e.g. > 65°C for 15 minutes) or pH extremes (< 6 or > 9).
- The OD260/OD280 ratio is approximately 1.8-1.9 and the OD260/OD230 ratio is 2.0-2.2.
- Contains no insoluble matter.
- Free of RNA contamination.
- No exposure to embedded fluorescent dyes or UV radiation. SYBR dye does not damage DNA. Avoid the use of ethidium bromide.
- Contains no denaturants (such as guanidine salts or phenol), detergents (such as SDS or Triton X100), or chelating agents.
- Does not contain origin from original organisms/tissues (heme, humic acid, polyphenols, etc.)
- DNA size should exceed 50kb as assessed by pulsed-field gel or equivalent.
Amount of DNA Required
The amount of DNA required for sequencing depends largely on the platform and the purpose of the study. For example, the Oxford Nanopore Technology platform may require microgram amounts of DNA, especially if deep sequencing coverage is the goal. It is also important to consider that some of the initial quantities may be lost during the library preparation step, thus requiring starting with a higher quantity.
DNA Extraction Protocol
(1) Sample preparation: Start with fresh or quick-frozen plant material. The choice of tissue affects the quality and quantity of extracted DNA.
(2) Cell lysis: Lysing cells is essential to release DNA. This process is aided by the use of pre-warmed SDS lysis buffer supplemented with β-mercaptoethanol, ensuring complete cell lysis and DNA release.
(3) Protein and polysaccharide precipitation: After lysis, the addition of potassium acetate helps precipitate proteins and polysaccharides, which can interfere with DNA quality.
(4) Phenol: Chloroform Extraction: This step helps purify the DNA and remove any remaining proteins or contaminants.
(5) Magnetic bead purification: Using carboxylated magnetic beads (such as Sera-Mag SpeedBeadsTM), short DNA fragments can be effectively removed to enrich high molecular weight DNA in the sample.
(6) Quality control: After DNA is extracted, its quality must be verified using spectrophotometry, electrophoresis, and DNA integrity values.
DNA Quality Assessment
DNA integrity: essential for accurate sequencing
DNA integrity is at the forefront of quality assessment. The presence of degraded or fragmented DNA can severely impact the results of long-read sequencing. Methods such as pulsed-field gel electrophoresis (PFGE) and Tapestation were used to determine the integrity of high molecular weight DNA. These technologies provide a comprehensive overview of the DNA size range, with tools such as Agilent Femto Pulse, Biorad Chef Mapper, and Sage Pippin Pulse providing a broad range of sizes, from as low as 0Kb to as high as 10Mb.
Purity: Ensures DNA is free from contamination
DNA purity is critical to eliminating the risk of confounding variables in sequencing results. Contaminants can interfere with sequencing reactions, causing massive inaccuracies. Devices such as the NanoDrop® Spectrophotometer can determine DNA purity using metrics such as the A260:A280 and A260:A230 ratios. For example, a low A260:A280 ratio indicates potential protein contamination. At the same time, deviations between readings from tools like NanoDrop and Qubit can be a red flag for serious contamination.
Concentration: Precise measurement for reliable sequencing
Accurate measurement of DNA concentration is critical for optimal sequencing results. Qubit® and Picogreen® are the primary tools in this field, providing double-stranded DNA (dsDNA) specificity. Ensuring the correct concentration is key to successful library preparation and downstream sequencing applications.
Reference
- Russo, Alessia, et al. "Low-input high-molecular-weight DNA extraction for long-read sequencing from plants of diverse families." Frontiers in Plant Science 13 (2022): 1494.
Related Services
PacBio SMRT Sequencing Technology
Oxford Nanopore Sequencing Technology
PacBio Pre-Made Library Sequencing
Oxford Nanopore Pre-Made Library Sequencing
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment