CD Genomics is one of the largest providers of long-read sequencing services. We continue to equip ourselves with the latest long-read sequencing platforms. We are offering long-read sequencing services from Pacific BioSciences (PacBio) and Oxford Nanopore Technology (ONT), providing researchers with a wide range of cutting-edge sequencing services to meet the specific needs of any project.
Long-read sequencing technologies offer the unique advantage of long-read length, which greatly enhances de novo assembly, mapping certainty, detection of structural variants, identification of transcriptional isoform, and so forth. Moreover, long-read sequencing methods enable direct detection of native DNA or RNA molecules. The use of these advanced sequencing methods has risen dramatically as sequencing costs have decreased and accuracy and throughput have continued to advance.
Long reads from PacBio SMRT and Oxford Nanopore sequencing technologies differ in length, throughput, and accuracy due to sequencing chemistry and sample preparation. These diverse data types have been applied to specific applications. For example, Oxford Nanopore sequencing takes full advantage of the ultra-long read length to easily span these complex structures in the microbial genome. While PacBio SMRT sequencing with high accuracy (HiFi data) is more suitable for studying gene regulation and genome complexity.


We are dedicated to providing outstanding customer service and being reachable at all times.

The PacBio Sequel II and IIe Systems
Sequel II and Sequel IIe both generate up to 8x more sequencing data per SMRT-cell and belong to the third-generation PacBio sequencers. The principles of their single-molecule sequencing chemistry are unchanged and their sequence coverage and sequence errors do not show any detectable sequence-specific bias. These PacBio Sequel sequencers enable HiFi reads, which is changing genomics.
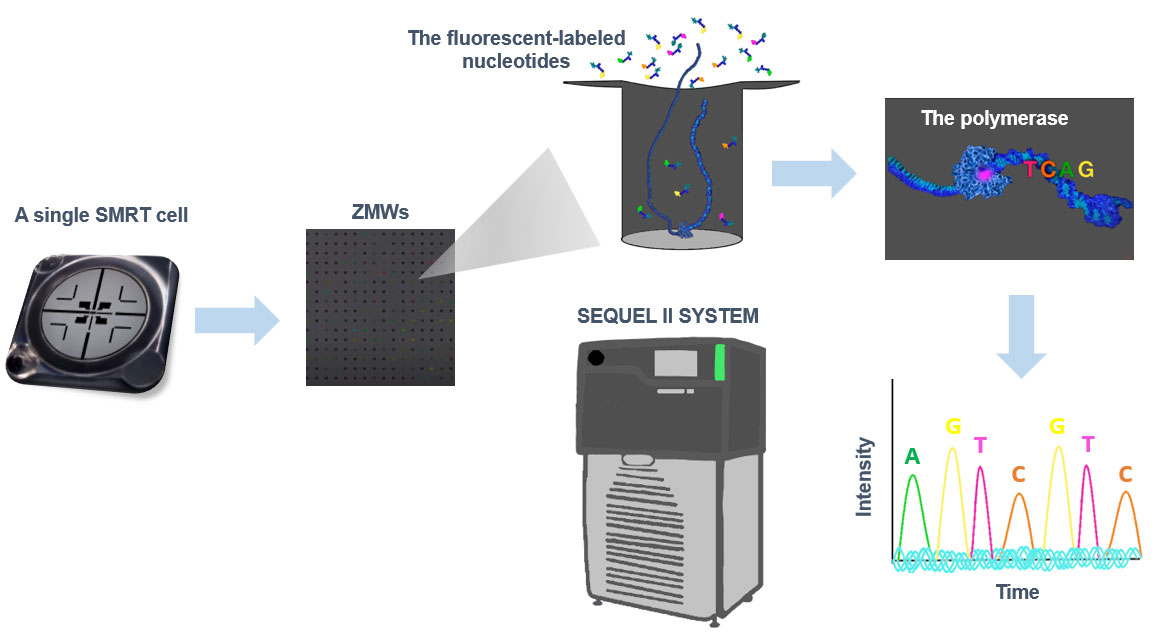
Applications of PacBio SMRT Sequencing
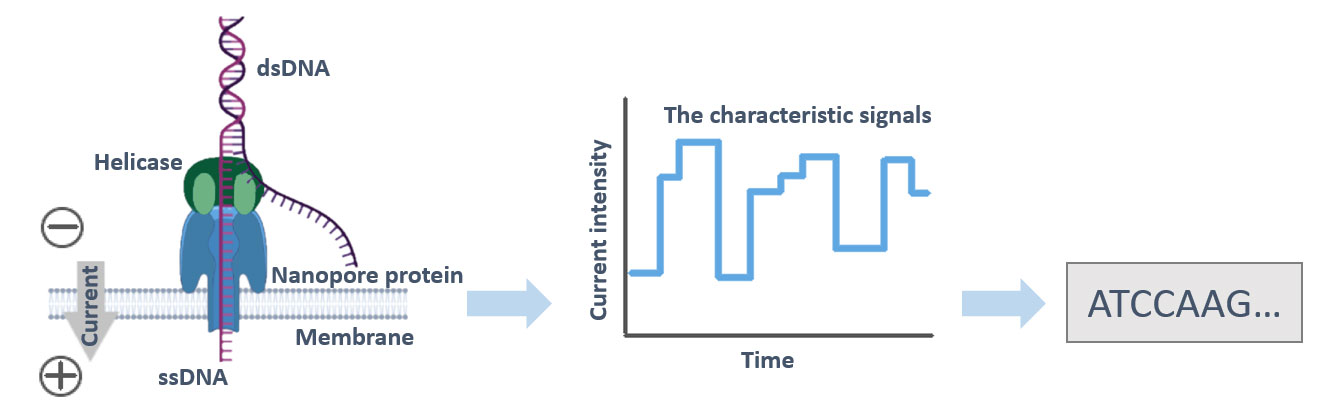

ONT Nanopore Sequencing
Nanopore sequencing with ONT systems can yield huge read lengths due to its unique pore chemistry, which allows molecules to be transferred through the nanopore regardless of length.
Outstanding Advantages of ONT
Long reads with
no amplification
The long read length of over 150 Kbp provides a more comprehensive display of the sequence information of the target gene and facilitates the identification of duplicated gene fragments and structural variants.
Direct detection of
target nucleic acids
Enable direct sequencing of epigenetic modifications and RNA.
Real-time access to
sequence information
Direct detection of the native strand of interest saves a lot of time. Sequencing signals are collected directly when the target nucleic acid passes through the nanopore, and sequencing results can be output in real-time.
Simple library
preparation
No PCR amplification, no fluorescent molecular markers, no optical imaging, extremely fast library preparation process.
ONT Long and Ultra-Long Reads
With the introduction of the Nanopore sequencing system, we have built a robust, scalable platform consisting of the MinION, GridION, and PromethION systems. These systems can now generate two main types of long reads, standard long reads (10-100kb) and specialized ultra-long reads (>100kb) with different lengths, accuracy, and throughput for specific applications.
ONT Nanopore Sequencing has many applications, including,
- DNA sequencing, including long-read and ultra-long-read DNA sequencing.
- Full-length cDNA sequencing.
- Direct RNA sequencing.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment