

We are dedicated to providing outstanding customer service and being reachable at all times.
Full-Length Transcriptome Profiling Combined with Long- and Short-Read Sequencing
CD Genomics provides comprehensive transcriptome profiling services using both short-read (Illumina) and long-read RNA sequencing technologies (PacBio Iso-Seq and ONT nanopore RNA sequencing). Our integrated analysis will facilitate the discoveries of novel transcripts and genes, identification of alternative splicing, and detection of allele-specific expression.
Methodologies for Transcriptome Profiling
Transcriptomics is the study of the transcript catalog of a cell, tissue, or organism under specific conditions (physiological condition, developmental stage, and external environment), involving protein-coding messenger RNA (mRNA) and non-coding RNA (ncRNA), such as transfer RNA (tRNA), ribosomal RNA (rRNA), and among others. Due to the complexity of the transcriptome, transcriptome analysis relies heavily on the availability of high-throughput tools. RNA sequencing using short-read sequencing (Illumina) is currently the standard and widely used approach for transcript profiling. This method is particularly suitable for large-scale, high-throughput studies. While the emerging long-read RNA-seq technologies, such as PacBio SMRT sequencing and nanopore cDNA/RNA sequencing, offer new ways to investigate the transcriptome and its function.
Methodologies for Transcriptome Profiling |
|||||
| Representative Technologies | Read lengths | Advantages | Disavantages | Main Applications | |
| Short-read sequencing technologies | Illumina | ~300bp | -High throughput. -Cost-efficient. -Effective in quantifying transcript abundance. |
-Read length limitations. -Need to amplify or fragment. -Complex splicing. -Amplification bias. |
-Identification and quantification of transcripts. -Detection of gene expression changes. |
| Thermo Fisher (Ion Torrent) | ~600bp | ||||
| MGI (DNBSEQ) | ~400bp | ||||
| Long-read sequencing technologies | Pacific Biosciences (PacBio) | ~15 kb | -Long-read length exceeds the lengths of most transcripts. -Without the need to amplify or fragment. -Direct cDNA/RNA sequencing(ONT). |
-Relatively high cost. -Relatively high error rate. -Relatively lower throughput. |
-Characterization of full-length transcript. -Systematical characterization of the complexity of transcript isoforms. -Identification of gene fusions. -Identification of RNA base modifications. (ONT) -Analysis of poly-A tail lengths. (ONT) |
| Oxford Nanopore Technologies (ONT) | > 30 kb | ||||
Outstanding Advantages of Our Services
- The short reads correct the long reads to produce more accurate transcriptome information.
- The combination of short-read and long-read RNA-seq enables systematical characterization of the complexity of transcript isoforms.
- The combination of short-read and long-read RNA-seq enables more comprehensive transcriptome analysis.
Workflow of Our Service

Sample Requirements
-Sample type: Blood, tissues, cells, RNA samples (total RNA or poly-A+ RNA), and more.
-RNA amount ≥ 2 μg, RNA concentration ≥ 50 ng/μL, OD260/280=1.8~2.0.
-All RNA samples are validated for purity and quantity.
Sequencing platforms
-Illumina NovaSeq and HiSeq platform.
-PacBio Sequel II sequencing system.
-Nanopore PromethION sequencing platform.
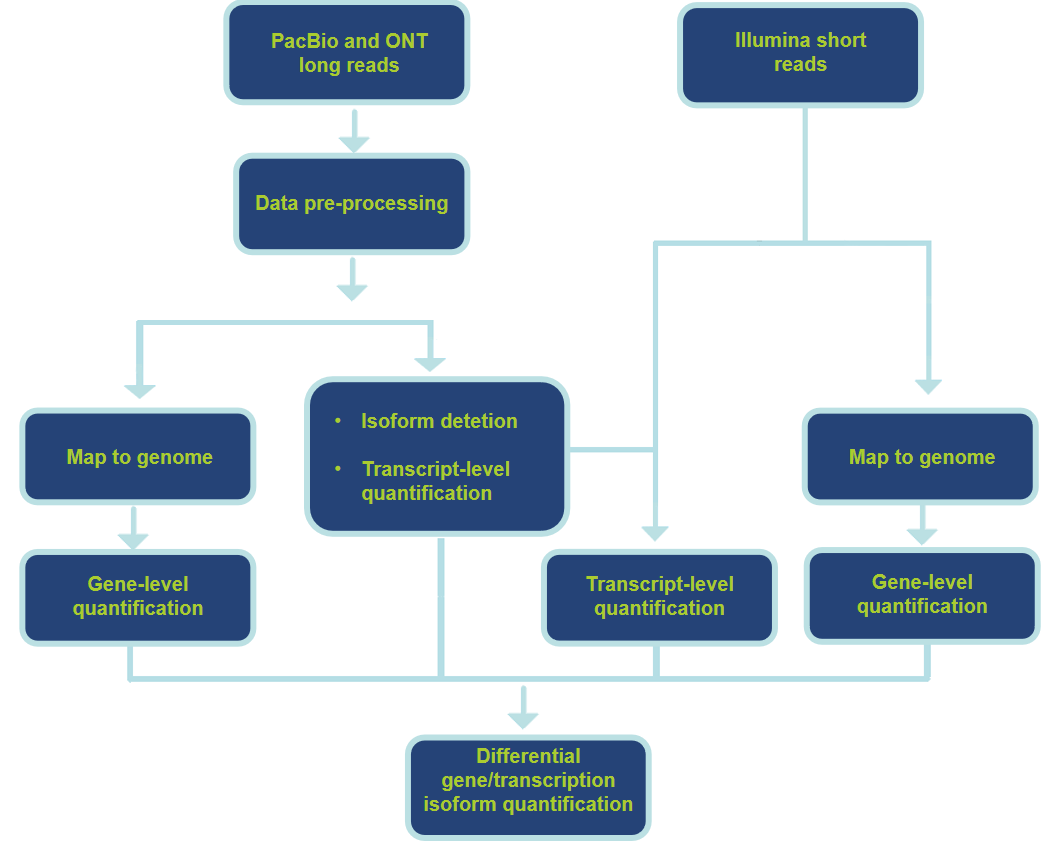
Analysis Pipeline and Contents
Why Do You Choose Us?
- Advanced instruments and highly experienced analysts.
- Proper methods of sample preparation and RNA extraction.
- Customized service to customer satisfaction.
- Complete analytical study reports.
- Our customer service representatives are available 24 hours a day from Monday to Friday.
With multiple specialists and years of experience in RNA-seq, we guarantee you high-quality data and integrated bioinformatics analyses. If you are interested in our short-read and long-read RNA sequencing, please feel free to contact us. We are looking forward to working on your next project.
References
- Dong, X., et al. (2021). "The long and the short of it: unlocking nanopore long-read RNA sequencing data with short-read differential expression analysis tools." NAR genomics and bioinformatics, 3(2), lqab028.
- Huang, K. K., et al. (2021). "Long-read transcriptome sequencing reveals abundant promoter diversity in distinct molecular subtypes of gastric cancer." Genome biology, 22(1), 1-24.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment