Biomarkers are objective indicators of an individual's health status that can be measured precisely and consistently. In order to better understand disease pathogenesis and related changes, and promote the development of disease treatments, robust and validated biomarkers are needed. Conventional methods provide far less perspective from biomarker genomics than our next-generation sequencing (NGS), genotyping, and knowledge.
Biomarker Genomics in Gut Microbiome
Recent breakthroughs have increased biomarker discovery and profiling in NGS, microarray, and bioinformatics. Scientists can use biomarker genomics to create enough information to extend the human genome and answer a variety of questions at the same time. Different techniques, such as DNA, RNA, and methylation-based profiling on a global scale or at specified areas, are used in biomarker genomics. This enables scientists to identify the known and new biomarkers in a high-throughput and precise manner and assist the clinic from drug exploration to decision-making. In customized medicine biomarker genomics is also utilized to anticipate medical diagnosis and treatment based on a patient's genetic information.
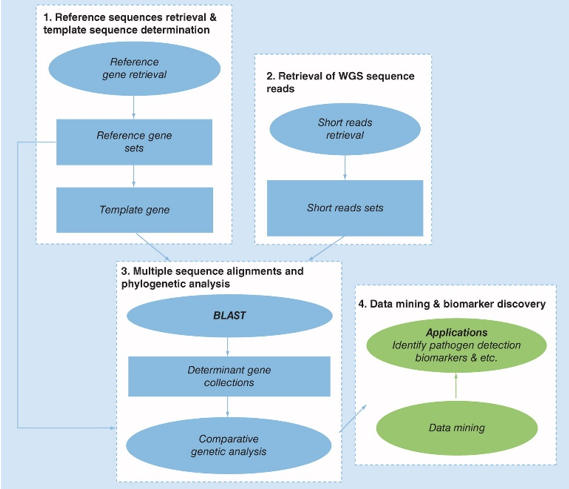
Figure 1. Schematic representation of the proposed bioinformatics pipeline for biomarker discovery by WGS. (Zhao, 2015)
About Our WGS-Based Biomarker Identification Service
CD Genomics is a well-known company that focuses on multi-omics research. We have vast knowledge associated with all sorts of issues you may have experienced in your studies, integrated with cutting-edge methods and provided by well-experienced researchers. As the investigation on microbiota biomarkers is rapidly expanding due to the excellent progress of multi-omics technologies and sources, we can give powerful tools and the most legitimate solutions to support your studies on intestinal microbiota and biomarkers, emphasizing the important information that gut microbiota biomarkers could give to show light upon researches into physiological processes, metabolic pathway, pathogenesis.
Whole-genome sequencing is a single-time determination of the full DNA sequence of the genome of an organism, involving both chromosomal DNA and DNA stored in mitochondria and chloroplasts. With increased accuracy, whole-genome resequencing can detect DNA biomarkers like single nucleotide polymorphisms (SNPs), insertions and deletions (indels), structure variations (SVs), copy number variations (CNVs), and other genetic alterations in the sequenced species. It also offers a once-in-a-lifetime opportunity to characterize polymorphic variations in a population, revealing the underlying processes of species origin, development, growth, and evolution in great detail.
We can offer thorough biomarker assessment from all genetic levels, transcription levels, expression levels, and metabolic levels, and assist you to determine particular biomarkers and explore single or multiple biomarkers that have not been defined yet, because our expertise includes the complete spectrum of multi-omics, such as metagenomics, metatranscriptomics, metaproteomics, and metabolomics.
Our Advantages
- Single base-pair resolution
- Classification of de novo sequencing and genome-wide mutation
- Evolution and phylogenetic analysis of the population
- Drug exploration and development, as well as customized medicine, are all areas in which researchers are working.
Workflow and Bioinformatics Analysis
| Analysis Contents | Details |
| De novo Assembly | Creating genomes from a great amount of (short or long) DNA fragments with no prior knowledge of their appropriate sequence or order. |
| Reference Genome Mapping | Discover where reads emerge from in the genome and then either call variants or build a consensus sequence for the sequence of aligned reads when these reads are synchronized. |
| SNP/InDel/SV/CNV Calling | Single nucleotide polymorphisms, insertions/deletions, structural variations, and copy number variations can all be found using this method. |
| Annotation | Classifying gene locations and all coding areas in a genome, as well as identifying what those genes do. |
| Pathway Analysis | Taking rates of gene expression and leveraging current information of the specified organism to define the underlying biological processes and mechanisms. |
In gut microbiota studies, CD Genomics is dedicated to giving the highest level of solutions. We'll recommend the best programs based on your specimen and research objectives. And we would be more than pleased to support your studies into work requiring intestinal microbiota and to advertise human health and well-being. Please feel free to approach us to know out more options for the exploration of gut microbiota and biomarkers.
References
1. Torres J, Petralia F, Sato T, et al. Serum biomarkers identify patients who will develop inflammatory bowel diseases up to 5 years before diagnosis. Gastroenterology. 2020, 159(1).
2. Zhao W, Chen JJ, Foley S, et al. Biomarker identification from next-generation sequencing data for pathogen bacteria characterization and surveillance. Biomarkers in medicine. 2015, 9(11).
3. Iskandar HN, Ciorba MA. Biomarkers in inflammatory bowel disease: current practices and recent advances. Translational Research. 2012, 159(4).
*For Research Use Only. Not for use in diagnostic procedures.