The human gastrointestinal tract is colonized by a vast array of microbes, which form communities of bacteria, viruses and microbial eukaryotes that are specific to people of different ages, diet habits, and health conditions. Gut microbiota plays a pivotal role in human health and fitness. Metaproteomics is the large-scale identification and quantification of proteins in order to study the expression and function of proteins from microbial communities, is a versatile tool to provide insights into the phenotypes of microorganisms on the protein level.
Two-dimensional liquid chromatography (2D-LC) is the most widely used multidimensional liquid chromatographic technique in metaproteomic research and is compatible with the separation of proteins with large differences in molecular size, low abundance proteins, and hydrophobic proteins. 2D-LC involves connecting two different chromatographic columns in sequence, in which the injected sample is separated by passing through two different separation stages. 2D-LC can be used to address problems of complex naturally occurring mixtures, biomarkers for various biological conditions, (meta)proteomics and metabolomics. When used in combination with mass spectrometry, 2D-LC provides a huge amount of qualitative and quantitative data, which significantly improves the depth of detecting and plays a key role in the separation of complex and difficult-to-separate materials in the fields of proteomics and metaproteomics. However, these improved separations typically came at the cost of long analysis times (for example, several hours to days) and thus 2D-LC was really a niche technique limited to a small fraction of all liquid phase separations.
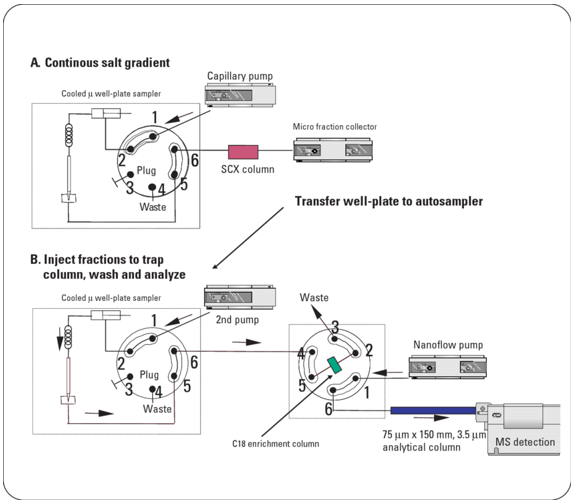
Figure 1. Workflow for proteome identification by off-line 2D LC/MS (Pieper 2013)
CD Genomics, as a preeminent corporation that has many years of experience in proteomics. We present the state-of-the-art 2D-LC technique for targeted and untargeted metaproteomic analysis, we also offer alternative strategies for 2D-LC data analysis:
- Treat 2D-LC data as a series of 1D chromatograms each of which is analyzed in a more or less conventional fashion.
- Turn 2D-LC data into a 2D image at the outset and then treat it by various techniques of image and feature analysis.
- The higher-order structures of the data (e.g. 2D-LC plus MS) are treated analyzed by chemometric methods such as parallel factor analysis (PARAFAC), the generalized rank annihilation method (GRAM), or multivariate curve resolution-alternating least squares (MCR-ALS).
We dedicate to providing the highest level of service in gut microbiota research. We will offer the most preferred strategies according to your sample and research purpose. To find out more about our 2D-LC services, please feel free to contact us.
Reference
1. Pieper R; et al. Proteomics and Metaproteomics. In: Nelson K. (eds) Encyclopedia of Metagenomics. 2013.
*For Research Use Only. Not for use in diagnostic procedures.