Piwi-interacting RNAs (piRNAs), generally 21–36 nucleotides in length, are the largest class of small non-coding RNA molecules expressed in animal cells. piRNAs are mostly created from loci that function as transposon traps and provide an RNA-mediated adaptive immunity against transposon expansions and invasions, and they can also be involved in the regulation of other genetic elements in germline cells. Like other small RNAs, piRNAs are thought to be involved in gene silencing, specifically the silencing of transposons. Other functions include preserving genomic integrity, remodeling euchromatin and epigenetic programming, regulating translation, regulating target mRNAs, modulating mRNA stability, developmental regulation, etc.
The pathway of piRNAs is less well understood than other small RNAs like miRNA and siRNA. The wide variation in piRNA sequences and piwi function over species contributes to the difficulty in establishing the functionality of piRNAs, and individual piRNAs are not conservative, which makes the deductions of their functions more challenging. Though some studies showed that piRNAs have certain functions on the biogenesis, germline, and brain; abnormal expression of piRNAs is associated with many kinds of diseases including cancer. The findings remain preliminary, that piRNAs may or may not have direct or indirect interactions with gut microbiota or gut-brain axis still remains an intriguing topic to research.
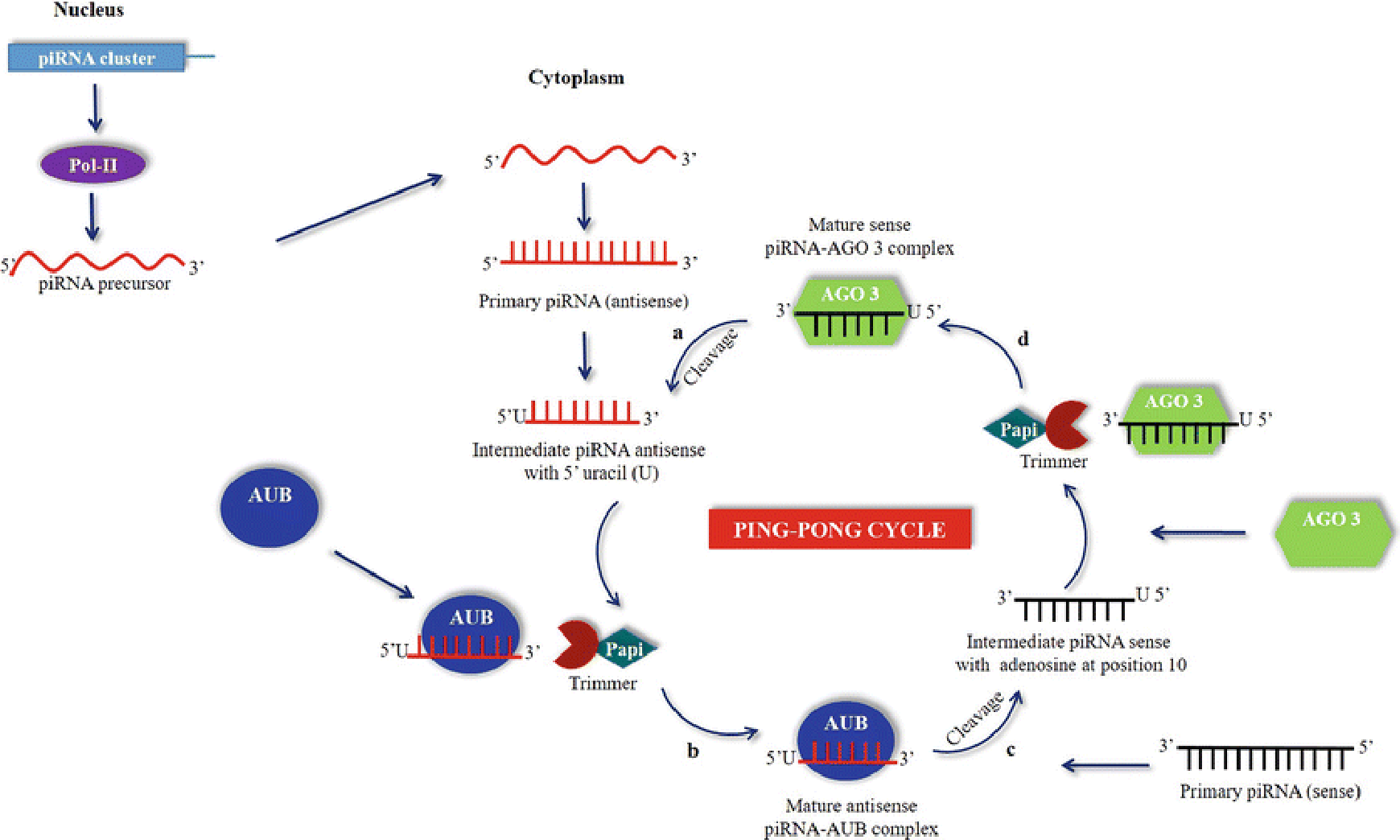
Figure 1. Role of Piwi-interacting RNA (piRNA) in epigenetic programming (Luteijn 2013)
CD Genomics provides approaches to piRNA profiling and mechanism uncovering including piRNA sequencing based on next-generation sequencing and piRNA microarray.
piRNA Sequencing
We provide piRNA profiling service by using next-generation sequencing or massively parallel high-throughput DNA sequencing, allows for the examination of tissue-specific expression patterns, disease associations, or isoforms of piRNAs, and to discover previously uncharacterized piRNAs. With advanced technology and experienced experts, we possess the full ability to:
◆ Obtain piRNA abundance levels from sequence reads
◆ Discover novel piRNAs
◆ Determine the differentially expressed piRNA
◆ Determine the piRNA associated gene silencing targets
piRNA microarray
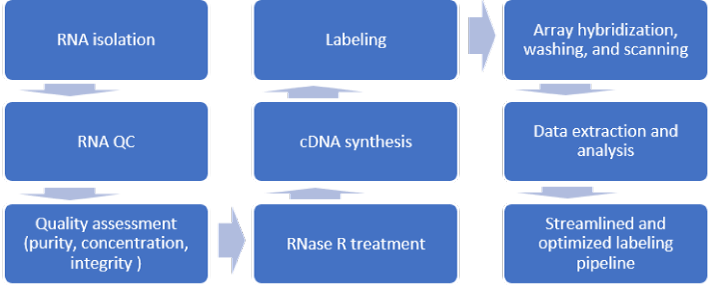
Microarray is a powerful high-throughput tool that is capable of conducting a genome-wide analysis of piRNA mechanisms, detecting multiple samples in parallel and monitoring the expression of thousands of small noncoding RNAs in a single run. We provide one-stop service of piRNA microarray analysis, the major protocol includes piRNA oligo probe design, array fabrication and piRNA target preparation, target-probe hybridization on array, signal detection and data analysis. The process can be standardized and allows the reproducible profiling of piRNA.
Our piRNA profiling service would assist you to excavate more abundant information from your research. To find out more about the piRNA sequencing and microarray services, please feel free to contact us.
Reference
1. Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nature Reviews Genetics. 2013, 14(8): 523-34.
*For Research Use Only. Not for use in diagnostic procedures.