CD Genomics offers microbial 16S/18S/ITS amplicon sequencing services, metagenomic sequencing services and professional bioinformatics analysis, which can quickly and accurately identify the species composition and diversity of microorganisms in the sample, and obtain a variety of biological information such as community evolutionary relationship, species structure and community function.
Microbial 16S amplicon sequencing
16S amplicon sequencing uses suitable universal primers to amplify microorganisms' 16S rDNA hypervariable regions or functional genes. It is a high-throughput sequencing technology that is used to detect the sequence variations and abundant information of PCR products, analyze the diversity and distribution of microbial communities in the environment, and reveal the types, relative abundance, and evolutionary relationships of microorganisms in environmental samples.
18S rDNA or ITS (Internal Transcribed Spacer) is widely used in the classification and identification of eukaryote and fungi. 18S rDNA is more suitable for the classification of elements above species level in phylogenetic research. ITS sequences are located between 18S, 5.8S and 28S rRNA genes of fungi, which are ITS 1 and ITS 2, respectively. ITS belongs to a moderately conservative region, and it can be used to study species and classification levels below species.
- Bioinformatics analysis
OTU clustering and analysis
Species classification
Single sample diversity analysis (alpha diversity)
Comparative analysis of diversity among samples (beta diversity)
Species difference analysis
Function analysis: KEGG function prediction, COG function prediction
Association analysis and model prediction
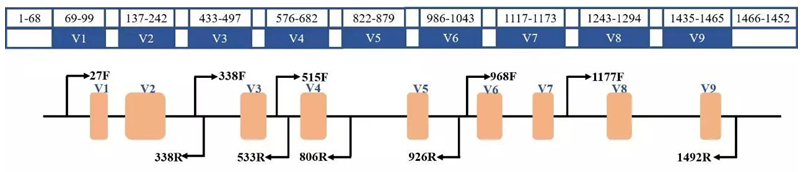
For 16S sequencing, any one or several highly variable regions, despite their high variability, may be very similar for some species. The specific sequence fragments that can distinguish them may not be in the amplified region. Therefore, the full-length variable region sequence can be selected for determination, which covers a wide range and can accurately identify the species level.
Microbial 16S metagenomic sequencing
Metagenomics takes the entire microbial community in a specific environment as the research object. There is no need to separate and culture, and DNA from environmental samples can be directly extracted for sequencing. The 16S metagenomics can be used to study the community structure, species classification, system evolution, gene function, and metabolic network of environmental microorganisms.
- Bioinformatics analysis
Gene prediction and gene set construction
Genetic analysis: Venn diagram and difference analysis
Gene set function annotation: Eggnog, KEGG, CARD, COG, CAZy, Swiss-prot, NR, etc.
Species analysis
Functional analysis
Diversity analysis
Network interaction
Similarity analysis
Association analysis and model prediction
How to choose?
| Amplicon sequencing | Metagenomic sequencing | |
| Principles of sequencing | The extracted microbial genomic DNA is amplified by PCR on a certain or several hypervariable region sequences (V4 region or V3-V4 region), then the library is constructed and sequenced. | Microbial genomic DNA is randomly separated into small fragments of 300-500bp. Add sequencing adapters at both ends of the fragments and perform sequencing. |
| Depth of species identification | Many sequences obtained by sequencing are not annotated at the level of species. | Can identify microorganisms to the species level or even the strain level. |
| Research purposes | Mainly study the species compositon of the community, the evolutionary relationship between species and the diversity of the community. | Based on 16S sequencing analysis, in-depth research on genes and functions (GO, Pathway, etc.) can also be carried out. |
CD Genomics can provide you with microbial 16s amplicon sequencing, metagenomic sequencing, and high-quality result reports. If you have any questions about the above services or consult other microbiome analysis services, please contact us.
*For Research Use Only. Not for use in diagnostic procedures.