CD Genomics provides microbial metagenomics analysis for biology, environment, medicine and other fields. We use high-throughput next-generation sequencing (NGS) technology to analyze microbial diversity, population structure, evolutionary relationships, etc., using the microbial population genome in a specific environment as the research object. Explore the functional characteristics of microbial populations, their mutual cooperation and the relationship with the environment, and explore the potential biological significance.
Advantages of our microbial metagenomics service
1. High-throughput sequencing platform: We have a variety of high-throughput sequencing platforms such as Illumina HiSeq 2500 and HiSeq 4000.
2. Low sample demand: The recommended sample size for the conventional metagenomic library is 500ng or more. For samples that are difficult to obtain, you can choose to build a micro-database, and the sample size can be as low as a few ng.
3. Strict quality control process: Optimized data quality control, including filtering and matching sequences with low quality, non-specific amplification, low coverage and low complexity, so as to quickly and accurately obtain microbial information and abundance information in samples and maximize data quality.
4. Comprehensive bioinformatics analysis: species analysis, gene prediction and analysis, diversity and similarity analysis, function analysis, network interaction analysis, metabolic network, association analysis, etc.
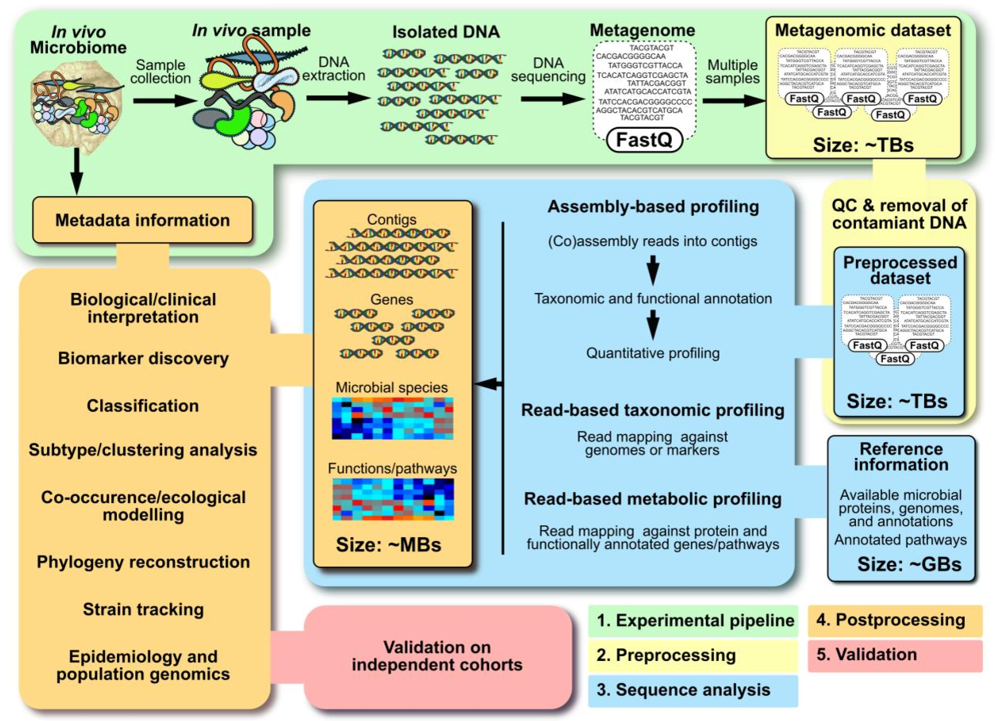
Summary of a metagenomics workflow (Quince et al., 2017)
Bioinformatics analysis
| Species analysis | Species composition analysis | |
| Species difference analysis | Venn diagram, Lefse analysis | |
| Similarity analysis | PCA analysis, PCoA analysis, Bray-Curtis distance, NMDS analysis, UPGMA analysis, cluster heat map, Anosim analysis | |
| Network interaction analysis | ||
| Functional Analysis | Differential functional analysis | LEFSE analysis, KEGG differential metabolic pathway analysis, KEGG enrichment analysis |
| Similarity analysis | Heat map clustering | |
| Correlation analysis and model prediction | RDA/CCA analysis (need to provide environmental factor information) | |
| PERMANOVA/Adonis permutation multivariate analysis of variance | ||
| Correlation coefficient Pearson/Spearman analysis (Spearman correlation coefficient analysis requires phenotypic information) | ||
| Random forest classification, tree classification effect analysis (do not do this analysis if the sample size is small) | ||
| ROC curve (do not do this analysis if the sample size is small) | ||
| Procrustes and Mantel test analysis (environmental factors or phenotypic information are required) | ||
| Circos diagram |
Services can be applied to the following areas:
1. Medical field: metabolic diseases, tumor cancer, intestinal flora metabolism and diseases, etc.
2. Livestock field: intestines, rumen (such as methanogenic bacteria) and animal health/nutrition digestion research, etc.
3. Agricultural field: the interaction of rhizosphere microorganisms and plants, agricultural farming/fertilization treatment and soil microbial community, etc.
4. Environmental field: haze treatment, sewage treatment, petroleum degradation, acid mineral water treatment and marine environment, etc.
5. Bioenergy: development of strains with special functions, gene mining, and engineering bacteria.
6. Special extreme environments: research on microbial groups under extreme environmental conditions
Sample requirements:
1. Environmental samples: soil 5-10 g, sludge/sediment 5-10 g, water body-filter membrane to filter to visible cover, feces 3-5 g, blood 10 mL.
2. DNA: total amount ≥ 1 μg, concentration ≥ 10 ng/μL, OD260/280=1.8-2.0, and ensure that the DNA is free from degradation and pollution.
3. The sample number of the sample delivery tube must be clearly marked, and the tube port should be sealed with Parafilm.
4. Avoid repeated freezing and thawing during sample storage, and use dry ice for transportation when sending samples.
CD Genomics can provide you with microbial metagenomic analysis and deliver raw data and detailed results reports. If you have any questions about the above services or consult other microbiome analysis services, please contact us.
*For Research Use Only. Not for use in diagnostic procedures.