Protein is the direct embodiment of gene function. Metaproteomics refers to all the proteins expressed by environmental microorganisms at a specific moment. The research types of Metaproteomics are very diverse, such as microorganisms in active sludge, marine microorganisms, soil microorganisms, fermented food microorganisms, intestinal microorganisms, feces, mucosal cavities, etc. On the basis of microbial metagenomic data, further research on its corresponding protein composition and function will be able to deeply explore the abundance distribution and functional orientation of functional molecules in the microbial population.
A large number of studies have demonstrated the application of metaproteomics in studying the functional role of gut microbiota in health and disease. Metaproteomics can not only study the composition and metabolic changes of the gut microbiota in disease states, but also consider the dynamic changes of biochemical pathways related to the disease.
CD Genomics has a high-resolution tandem liquid chromatography mass spectrometry platform (LC-MS/MS), based on the label-free proteomics Shotgun technology to elucidate gut microbes. In addition, we can also implement one-stop, personalized data analysis to help realize data visualization.
Which samples are more suitable for metaproteomics?
1. The composition is complex and there are many interferences
2. A wide variety of microorganisms
3. Largely affected by external environmental factors (season, temperature, humidity, etc.)
4. Complicated spatial location
5. The existence of other living things, such as animals and plants
What information can you obtain?
- Get all the protein information from different species in the sample
- The composition and function of the microbial community in the sample can be analyzed to reveal the relationship between microorganisms and the environment
- Reveal the changing classification, function, and metabolic pathways in the sample microbiota
Technology Platform of Metaproteomics Analysis
- Electrospray tandem mass spectrometry (ESI-MS/MS)
- Nano liquid chromatography tandem mass spectrometry (LC-MS/MS)
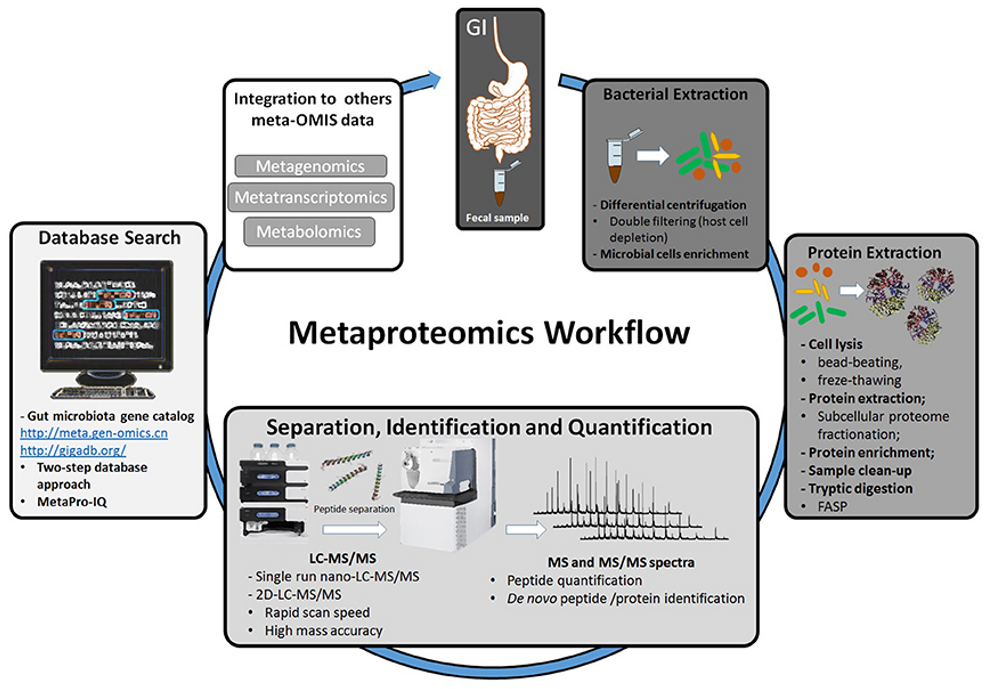
Workflow of metaproteomic analysis of fecal sample extracted from the gut (Petriz et al., 2017)
Applications of Metaproteomics
- Biomedicine: Research on the relationship between intestinal flora and major diseases.
- Microbiology field: pathogenic mechanism, drug resistance mechanism, pathogen-host interaction research, etc.
- Clinical diagnosis: biomarker discovery, disease mechanism, disease classification, etc.
Bioinformatics Analysis
- Go function annotation
- KEGG pathway annotation
- COG functional classification annotation
- Statistical Analysis
- Functional classification annotation and enrichment analysis
- Cluster analysis of expression patterns
- Protein interaction network analysis
- Species abundance analysis
Reference
1. Petriz, Bernardo A., and Octávio L. Franco. "Metaproteomics as a complementary approach to gut microbiota in health and disease." Frontiers in chemistry 5 (2017): 4.
*For Research Use Only. Not for use in diagnostic procedures.