The genes of gut microbes that comprise the microbiota surpass human genes by more than 100-fold, with over 3 million bacterial genes in the gut on its own, the microbiome has been dubbed "another organ" or "our second genome." Anaerobes make up over 99 percent of the bacteria in the gut, and many of them haven't been extensively examined because they can't be cultivated outside of their hosts. Microbiome evaluation that recovers the full-scale genetic data of gut microbiota has provided useful, efficient, and thorough services, which benefit tremendously from techniques such as PacBio SMRT Sequencing. These techniques are shedding light on gut microbiota studies, allowing researchers to gain an overview of human gut microbial complexity across various hosts in the hopes of addressing health problems that people face.
Highly repetitive components in both eukaryotic and prokaryotic genomes make genome assembly complicated and comprehensive studies of repetitive sequences hard. Long-read sequencing generates reads of several or dozens of kilobases (kbs), enabling the resolution of huge structural characteristics by spanning complex or repetitive areas with a single continuous read. It can show where methylated bases happen, giving detailed data about DNA methyltransferases encoded by the genome, in addition to much longer and incredibly reliable DNA sequences from specific unamplified molecules. PacBio SMRT sequencing offers distinct benefits in de novo genomics, metagenomics, transcriptomics, and epigenetics research.
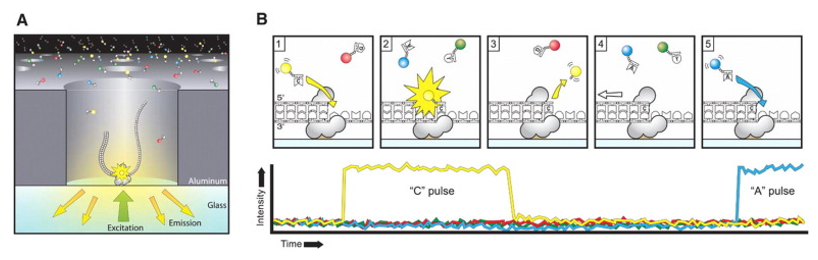
Figure 1. The principle of PacBio SMRT sequencing. (Rhoads, 2015)
About Our PacBio SMRT Sequencing Service
CD Genomics utilize the innovative PacBio SMRT devices (PacBio SR II and PacBio Sequel) are used for a variety of research projects, such as whole de novo genome assembly, full-length target sequencing, metagenomics research, full-length transcript sequencing, and genome-wide DNA modification evaluation. To guarantee confident and high certainty outcomes, our professionally trained expert team performs quality management after each procedure. De novo assembly, base modification identification, single-molecule consensus generation, transcript assessment, amplicon evaluation, and sequence orientation with variant identification are all part of our bioinformatics pipeline. More data mining options are accessible based on your requirements.
Our Advantages
- High Accuracy: Sequencing without systematic error accomplishes a consensus accuracy of >99.999 percent. Examine the advantages of highly precise long-read sequencing.
- Uniform Coverage: Because there is no bias based on GC material, you can sequence through areas that other techniques can't reach.
- Single-Molecule Resolution: Collecting sequence data from native DNA or RNA molecules allows for highly precise long reads with single-molecule precision of >99.9%.
- Base alterations are instantly identified during sequencing because there is no PCR amplification step.

Workflow and Bioinformatics Analysis
| Bioinformatics Analysis | Details |
| Gene Prediction | Recognises the gene-coding areas of genomic DNA. |
| Functional Annotation | KEGG, eggNOG, and CAZy are used to append biological information to gene or protein sequences. Comparing individual proteins in databases and allocating each one a specialized role. |
| Taxonomic Annotation | In a reference database, designates the taxonomy of a sequence. |
| Clustering | On the premise of a statistical distance structure, dividing observed datasets into a few subclasses or clusters. |
| PCA | The comparison of microbial metaproteomics between specimens and sample sets based on multiple factors. |
CD Genomics is dedicated to providing the highest level of service to facilitate gut microbiota research. The PacBio System is an extremely reliable and cheap metagenomics sequencing framework that should be used for complicated samples and low-diversity microbial populations. In long-read metagenomic sequencing, we will be your reliable partner. For more information and a detailed quote, please contact us.
References
1. Derakhshani H, Bernier SP, Marko VA, Surette MG. Completion of draft bacterial genomes by long-read sequencing of synthetic genomic pools. BMC genomics. 2020, 21(1).
2. Mahmoud M, Zywicki M, Twardowski T, Karlowski WM. Efficiency of PacBio long read correction by 2nd generation Illumina sequencing. Genomics. 2019, 111(1).
3. Sadowsky MJ, Staley C, Heiner C, et al. Analysis of gut microbiota–An ever changing landscape. Gut microbes. 2017, 8(3).
4. Rhoads A, Au KF. PacBio sequencing and its applications. Genomics, proteomics & bioinformatics. 2015, 13(5).
*For Research Use Only. Not for use in diagnostic procedures.