We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy
The introduction of next-generation sequencing has revolutionized molecular biology, and high-throughput sequencing systems are now commonly used in phylogenomic and metagenomic research. Though next-generation sequencing restricts the read-length of the process that ensures accuracy and fidelity, it does so while analyzing a significant number of amplified identical DNA strands. Because eukaryotic genomes include many repetitive areas, long DNA molecules must be split up into small parts, which could be a significant constraint of eukaryotic DNA sequencing. Third-generation sequencing techniques can sequence much longer reads than next-generation sequencing because they sequence single molecules instantly, resulting in a reformation in high-quality genome sequencing, enabling for unbiased, more precise classification and even higher taxonomic resolution than second-generation sequencing methodologies for gut microbiota research. The third generation of sequencing has surmounted the sophistication of short-read second-generation sequencing information assembly and the incapability to efficiently fix repeat sequences or huge genomic rearrangements; they all produce very long reads (1–100kb), which is unique from next-generation sequencing.
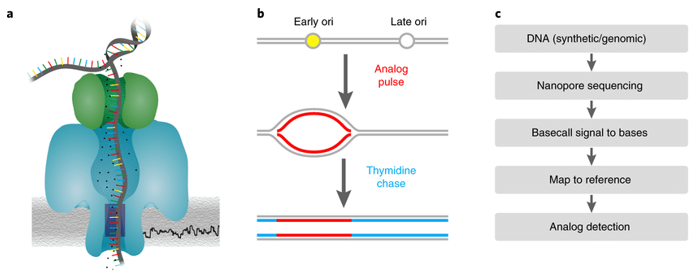
Figure 1. The principle and workflow of Nanopore sequencing. (Müller, 2019)
About Our Nanopore Long Read Sequencing Service
To assist your studies into gut microbiota, CD Genomics offers cutting-edge experimental protocols and algorithmic techniques for third-generation sequencing. Metagenomic sequencing, genome resequencing, genome de novo sequencing, transcriptome research, methylation identification, epigenomics experiments, disease-related variation detection, viral analysis, and other applications can all benefit from our third-generation sequencing services. We also provide services through third-generation sequencing and next-generation sequencing to obtain even more precise outcomes. Customized service will ensure the great quality of your subject and offer additional sparkly concepts for your studies, so please contact us to learn more about appropriate research strategies.
Our Advantages
- The mappable length (> 2 Mb) is slightly longer
- Since the nanopores can sequence numerous molecules, ONT MinION has a very large throughput
- The expense of generating ONT data is lesser. Because sequencing costs are a significant barrier to TGS application, ONT's relatively high throughput and low price make it interesting for a variety of applications.

Workflow and Bioinformatics Analysis
| Bioinformatics Analysis | Details |
| Remove redundant | Erase genetic redundancy, which occurs when two or more genes conduct the same purpose and inactivation of one of these genes has little or no effect on the biological phenotype. |
| Gene Functional Analysis | Recognising any functional element along a genome's DNA sequence, but with a focus on genes at first. |
| SNP Calling | Figuring bases that vary from the reference genome in genetic data, usually with a related confidence score or statistical evidence metric. |
| Gene Expression Analysis | Differential expression assessment and quantification of gene/transcript expression stages |
| Functional Enrichment Analysis | Categorizing gene or protein classes that are over-represented in a large number of genes or proteins and may be linked to disease phenotypes. |
| PPIs | Protein-protein interactions are detected and identified. |
CD Genomics is dedicated to providing the highest level of service to facilitate gut microbiota research. Our innovative ONT framework and services will be your best Full-Length Sequencing partner with their competence and dedication. For more details and a comprehensive quote, please do contact us.
References
1. Amarasinghe SL, Su S, Dong X, et al. Opportunities and challenges in long-read sequencing data analysis. Genome biology. 2020, 21(1).
2. Müller CA, Boemo MA, Spingardi P, et al. Capturing the dynamics of genome replication on individual ultra-long nanopore sequence reads. Nature methods. 2019, 16(5).
3. Kamathewatta KI, Bushell RN, Young ND, et al. Exploration of antibiotic resistance risks in a veterinary teaching hospital with Oxford Nanopore long read sequencing. PloS one. 2019, 14(5).
4. Nicholls SM, Quick JC, Tang S, Loman NJ. Ultra-deep, long-read nanopore sequencing of mock microbial community standards. Gigascience. 2019 May;8(5).
*For Research Use Only. Not for use in diagnostic procedures.
- SUITE 111, 17 Ramsey Road, Shirley, NY 11967, USA
- Tel: 1-631-338-8059
- Fax: 1-631-614-7828
- Email: info@cd-genomics.com