The fungal microbiome, identified as the mycobiome, is an element of the human microbiome that has not been widely investigated. Even though mycobiota makes up a small fraction of the whole human microbiome, researchers have been able to start to reveal the identity of our fungal commensals and assess their function in human health and illness using culture-independent methods using high-throughput sequencing methodologies.
In the guts of many mammals, including humans, mice, rats, pigs, and many ruminant and non-ruminant herbivores, fungi have been identified. In many human illnesses, including inflammatory bowel disease, graft versus host disease, Hirschsprung-associated enterocolitis, colorectal cancer, and developed progression of hepatitis B virus infections, fungi have been involved in the exacerbation. The declared appearance of fungi as a portion of the human microbiome, and their prospective position as contributors to health and disease, showcase the need for a more profound characterization of the healthy human microbiome. Understanding a healthy mycobiome will help recognize disease-contributing fungal organisms in studies and better classify health-important fungal bacterial relationships.
Fungus ITS Amplicon Sequencing
DNA Barcoding is one of the techniques regarded to be a new concept for identifying unidentified fungal samples quickly and accurately. The nuclear DNA (rDNA) area of the Internal Transcribed Spacer (ITS) has been the most sequenced area for identifying fungal taxonomy at the stage of species, and even within species. A highly polymorphic, non-coding region with sufficient taxonomic components is the ITS area. It is consequently able to differentiate sequences into levels of species. Although a few restrictions still hinder the ribosomal ITS as a universal barcode marker for fungi, the ITS will persist as the key choice for fungal detection. The search has already begun for substitute areas as a DNA marker to enhance fungal recognition, particularly in particular heredities.
About Our Service
CD Genomics provides a profile of multi-omics solutions as one of the largest suppliers of NGS offerings. 16S/18S/ITS amplicon sequencing is categorized by cost-efficiency, high speed, and practicality. In order to assist you to define and evaluate the microbial community, we provide comprehensive ITS amplicon sequencing service for gut fungus.
Our Advantages:
- ITS ribosomal RNA NGS offers a cost-effective technique for identifying strains that cannot be discovered using conventional ways
- Culture-free process that helps the entire microbial population to be analyzed within a specimen
- NGS gives the capacity to integrate multiple specimens in a sequencing run
- High sensitivity and accuracy
- Affordable price and professional team
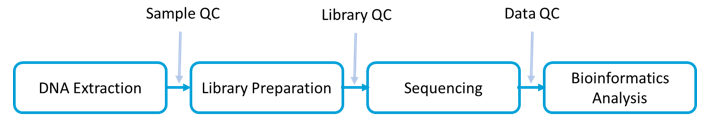
Workflow and Bioinformatics Analysis
| Bioinformatics Analysis | Details |
| Tags Assembly | For the reconstruction of the original sequence, aligning and integrating fragments from a lengthier DNA sequence |
| OTUs Clustering | Bacterial Taxa Approximation |
| Species Annotation | Classifying functional components along a genome sequence |
| Species Profiling Histogram | Assess in a specimen with regard to the species and their abundance. |
| Phylogenetic Tree | Display evolutionary interactions between different biological species or other entities based on commonalities and distinctions in their genetic features |
| Alpha Diversity | Mean species variety at a local scale in sites or ecosystems |
| Beta Diversity | Percentage between the variability of regional and local organisms |
CD Genomics provides gut fungus profiling services at high depth with high precision to meet the needs of studying population genetics and microbial profiles in the gastrointestinal tract, in order to research into biodiversity within and interaction between these microbes and human bodies. Our ITS amplicon sequencing-based fungus profiling protocol uses oligonucleotide probes designed to target and capture regions of interest – generally hypervariable regions of conserved genes or intergenic regions, followed by next-generation sequencing to meet the goals of efficient genetic variant identification and characterization by comparing against microbial databases and bioinformatic analysis. If you are interested in our amplicon sequencing service and want to find out more, feel free to contact us!
References:
1. Zhang YJ, Li S, Gan RY, et al. Impacts of gut bacteria on human health and diseases. International journal of molecular sciences. 2015, 16(4).
2. Kelly D, Conway S, Aminov R. Commensal gut bacteria: mechanisms of immune modulation. Trends in immunology. 2005, 26(6).
*For Research Use Only. Not for use in diagnostic procedures.