oxBS-Seq differentiates between 5mC and 5hmC. In this method, 5hmC is oxidized to 5fC with a selective chemical agent, while 5mC remains unchanged. Sodium bisulfite treatment of 5fC results in its deamination to uracil which, upon sequencing, is read as a thymidine. Deep sequencing of oxBS-treated DNA and sequence comparison of treated vs untreated libraries can identify 5hmC locations at single-base resolution. Now, CD Genomics provides you with a variety of oxBS-seq service to meet your research needs.
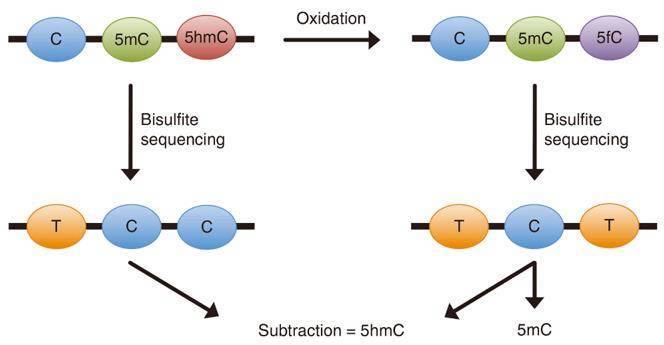 Figure 1. Schematic illustration of the principle behind oxBS-Seq (Booth M J.; et al. 2012)
Figure 1. Schematic illustration of the principle behind oxBS-Seq (Booth M J.; et al. 2012)
Specific oxBS-seq Approaches We Offer
Methods
- Process the genomic DNA to obtain a 10 kb fragment. Concentrate the fragmented DNA using a purification column.
- The fragmented and purified DNA was used for oxidation add bisulfite treatment. The 5-hydroxymethylcytosine oxidation was subjected to buffer exchange, denaturation, oxidation, bisulfite conversion, purification and qualitative evaluation.
- Finally, oxBS and BS-converted DNA were used to construct oxBS-seq and the corresponding BS-seq libraries. All double-stranded and single-stranded DNA was quantified on an Agilent 2100 Bioanalyzer prior to cluster generation and sequencing on the illumina platform, according to the manufacturer's protocol.
Our oxBS-seqservices are available for a wide range of sequencing applications:
- Whole genome oxidative methylation sequencing (oxWGBS)
- Simplified genomic oxidative methylation sequencing (oxRRBS)
- Target region target gene oxidative methylation sequencing (Target-oxBS)
Our bioinformatics analysis services include, but are not limited to:
- Sequencing data quality control
- Comparison with reference genome
- Genome-wide methylation and hydroxymethylation level distribution
- Correlation analysis
- Principal component analysis
- Differential methylation analysis and differential hydroxymethylation analysis
- Functional annotation using GO/KEGG analysis
- Differential expression level analysis of peak-associated genes
Services Advantages
- A completely new standard for DNA methylation detection.
- Single-base detection of DNA hydroxymethylation modifications.
- Multiplex quality control standards to detect oxidation efficiency and Bisulfite conversion rate.
- Low experimental preference and high reproducibility (R2>0.98).
- CD Genomics self-developed methylation-specific multiplex PCR primer design software.
Workflow
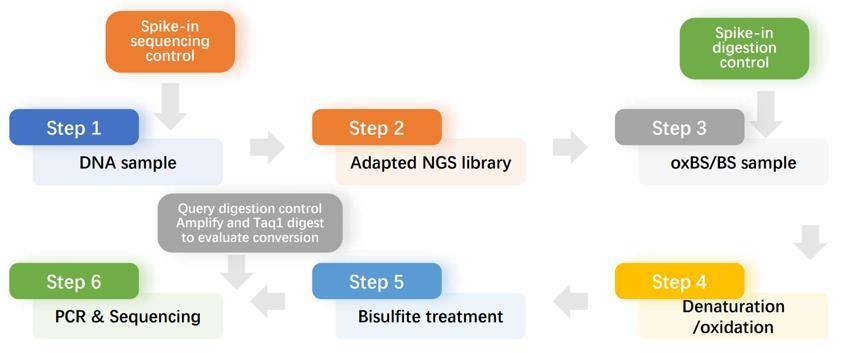
Sample Requirements
- Total DNA starting volume ≥ 5 µg (WGBS); ≥ 3 µg (RRBS)
- Sample concentration ≥ 50 ng/µl
- All DNA samples are verified for purity and quantity
Service Process

Deliverables
1. Related experimental results raw data
2. Experimental report
3. Data analysis
4. Image and result analysis
5. Bioinformatics analysis results
6. Details in oxBS-seq for your writing
Our Features
- CD Genomics has established a proven epigenomics sequencing platform for a variety of applications. We'll help you design the right oxBS-seq service to aid your scientific research.
- CD Genomics is a leading epigenomics sequencing technology company offering a trusted suite of services and solutions, especially with our extensive experience in oxBS-seq service.
Why Choose Us?
At CD Genomics, We offer innovative, flexible and scalable solutions that can meet the needs of our customers. We strive to meet this challenge as a global company that places a high priority on collaborative interactions, rapid delivery of solutions, and providing the highest level of quality. If you would like to know more about this service, please feel free to contact us.
Reference
- Booth M J.; et al. Quantitative Sequencing of 5-Methylcytosine and 5-Hydroxymethylcytosine at Single-Base Resolution. Science. 2012, 336(6083): 934-937.

 Figure 1. Schematic illustration of the principle behind oxBS-Seq (Booth M J.; et al. 2012)
Figure 1. Schematic illustration of the principle behind oxBS-Seq (Booth M J.; et al. 2012)
