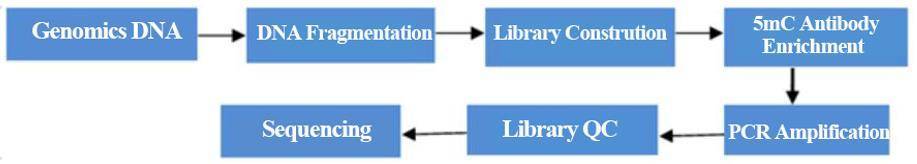
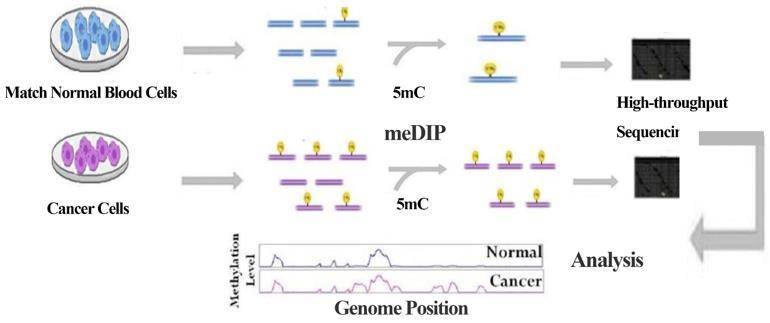
Methylated DNA immunoprecipitation sequencing (MeDIP-Seq) or DNA immunoprecipitation sequencing (DIP-Seq) is commonly used to study 5mC or 5hmC modifications. Specific antibodies can be used to study cytosine modifications. If 5mC-specific antibodies are used, methylated DNA is isolated from genomic DNA by immunoprecipitation. anti-5mC antibodies are incubated with fragmented genomic DNA and precipitated, followed by DNA purification and sequencing. Deep sequencing provides greater genomic coverage and represents the majority of immunoprecipitated methylated DNA. Now, CD Genomics provides you with a variety of MeDIP sequencingservice to meet your research needs.
Specific MeDIP Sequencing Approaches We Offer
We fragmented the extracted DNA, denatured it, ligated it to the junction and captured it using a 5mC antibody. The enriched methylated DNA fragments are then amplified and sequenced. With optimized experimental procedures and our highly specific 5mC antibody for enrichment of methylated DNA fragments, you can complete genome-wide DNA methylation studies with great efficiency and specificity at a lower cost.
Workflow
Services Advantages
- Genome-wide or any region of interest
- MeDIP Sequencing can target mC, mCG or hmC
- Nearly unbiased and hypothetical
- Epigenetic biomarker discovery
- Single nucleotide resolution and cost effectiveness
- Low input DNA requirements
Technology Strategies
Our Capabilities
- Our library builds are of high quality and have a very low failure rate.
- The whole process of quality control is traceable, with real-time tracking and timely feedback to ensure that the data are real and reliable.
- Our basic analysis is rich, and the presentation can be personalized.
- We provide advanced analysis services: professional bioinformatics analysis team, familiar with various analysis algorithms and software; according to the needs of customers, we provide a variety of personalized implementation solutions.
Sample Requirements
- We currently accept total DNA without degradation or RNA contamination or severe degradation
- Sample purity: OD 260/280 values should be between 1.8 and 2.0; RNA should be removed cleanly.
- Sample concentration: the low limit concentration should not be less than 50ng/ul.
- Total sample volume: not less than 1ug per sample.
- Sample solvent: dissolved in H20 or TE (pH 8.0).
- Sample transport: transport DNA at low temperature (-20°C); seal the tube with parafilm during transport to prevent contamination.
Service Process

Our Features
- We have accumulated many years of essence in the field of MeDIP sequencing service, helping customers to speed up project development and improve the overall success rate of the project.
- At CD Genomics, simply let us know your specific project needs. We will suggest the best strategy for you.
Deliverables
1. Related experimental results raw data
2. Experimental report
3. Data analysis
4. Image and result analysis
5. Bioinformatics Analysis
Applications
- Analysis of DNA methylation modification differences between samples
- Large sample size disease studies
Why Choose Us?
CD Genomics is a company that provides professional and comprehensive MeDIP sequencing services. We have years of experience to meet your specific project needs in using epigenomics research to add value to your research projects. CD Genomics can provide you with personalized solutions to help you thrive every step of the way around your interest in your workflow. If you would like to know more about this service, please feel free to contact us.



