Introduction to Bisulfite Sequencing PCR (BSP-Seq)
DNA methylation is one of the earliest discovered modification pathways, and methylation occurs in cytosines within CpG dinucleotides in eukaryotes. This process involves the conversion of cytosine at the 5' end of CpG dinucleotides to 5'-methylcytosine by DNA methyltransferases (DNMTs). Extensive research has shown that DNA methylation can cause changes in chromatin structure, DNA conformation, DNA stability, and the mode of interaction between DNA and proteins, thereby controlling gene expression. DNA methylation typically inhibits gene expression, while demethylation induces gene reactivation and expression. This mode of DNA modification allows for the regulation of gene expression without altering the gene sequence. Bisulfite Sequencing PCR (BSP) is a commonly used method for DNA methylation detection.
Principle of DNA Methylation BSP
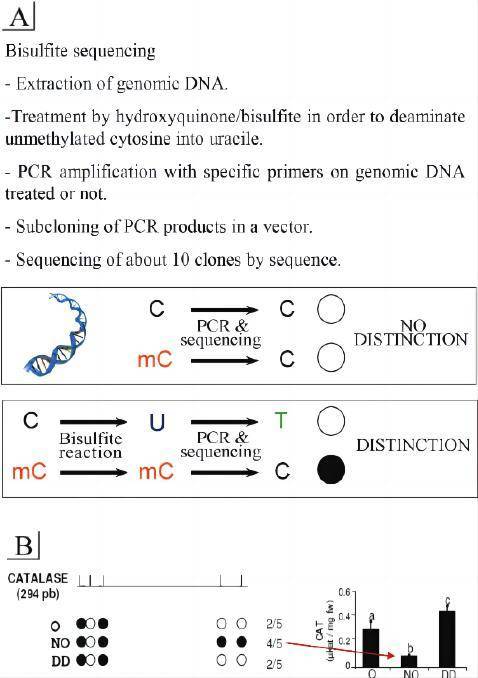
DNA methylation is present in most eukaryotes and generally occurs in the 5' UTR region of cytosines within CpG dinucleotides, playing a regulatory role. Approximately 80% of CpG cytosines are methylated, with 20% located in CpG islands in gene regulatory regions. The principle of Bisulfite Genomic Sequencing PCR (BSP) for methylation analysis involves treating genomic DNA with bisulfite, which converts unmethylated cytosines into uracil by deamination. In the subsequent PCR reaction, uracil is converted to thymine, while methylated cytosines remain unchanged and are preserved at the end of the reaction. The amplified PCR products are then subjected to TA cloning sequencing to analyze the specific methylation patterns. This method provides highly accurate results and serves as the gold standard for methylation detection.
Applications of Bisulfite Sequencing PCR
Early prediction, classification, grading, and prognosis assessment of tumors, as well as analysis of various diseases
Analysis of transcription levels: Methylation in promoter regions generally inhibits transcription
Study of regulatory mechanisms such as apoptosis
Bisulfite Sequencing PCR Service Content:
Design of specific primers based on the target gene sequence
Extraction and quality assessment of genomic DNA
Bisulfite modification of genomic DNA
Purification and recovery of modified DNA
BSP or qMSP experiments according to experimental requirements
Analysis of experimental results and provision of experimental reports to clients.
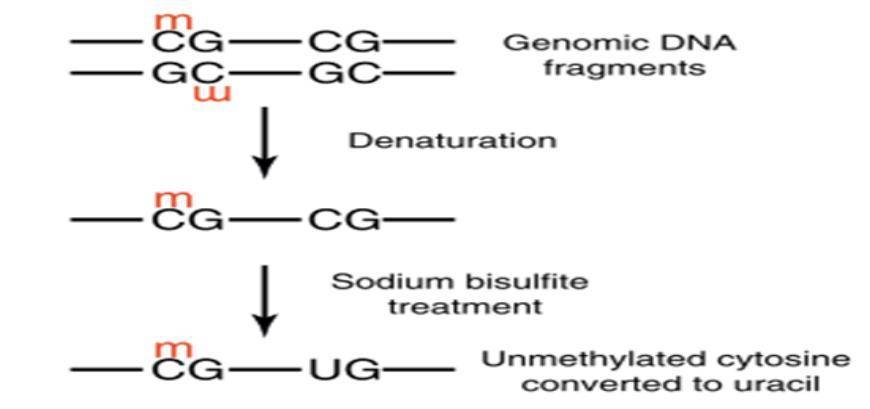 Principle of methylation analysis based on bisulfite conversion
Principle of methylation analysis based on bisulfite conversion
Features of Bisulfite Sequencing PCR Services:
Overcoming the limitations of Methylation-Specific PCR (MSP), which can only be used for qualitative research, our service allows for quantitative analysis.
High sensitivity, capable of detecting uneven distribution of 5-methylcytosine in the target DNA.
Free primer design provided by us when the customer provides the gene name or sequence.
Fast turnaround time.
Sample Requirements
(1) Customer-provided information: Gene name, species, and the specific methylation region or site of interest.
(2) Sample requirements: For cells, a minimum of 105 cells; for tissues, a minimum of 100 mg; for whole blood, a minimum of 0.5 ml; for genomic DNA, a minimum of 5 μg. It is recommended to ship all samples on dry ice to maintain a low temperature and ensure sample quality.
FAQ of BPS
Q :What are the common methods for DNA methylation analysis and what are their advantages and disadvantages?
| Common Methods | Advantages and Disadvantages |
|---|---|
| MSP method (Methylation-specific PCR, MSP) | Genomic DNA is treated with sodium bisulfite, and PCR is performed using primers specific to methylated and unmethylated sequences. Gel electrophoresis is used to detect the MSP amplification products. If amplification occurs with the primers specific to methylated DNA, it indicates methylation at that site; otherwise, it indicates the absence of methylation. |
| BSP method (Bisulfite sequencing PCR, BSP) | Genomic DNA is treated with sodium bisulfite, and BSP primers are designed for amplification. The PCR products are then cloned into a vector for sequencing to analyze the methylation status of CpG sites. |
| High-Resolution Melting (HRM) method | Genomic DNA is treated with sodium bisulfite. After PCR amplification, unmethylated cytosines are converted to thymine, while methylated cytosines remain unchanged. This results in changes in the melting temperature due to alterations in the GC content, which can be analyzed. |
Q: What are the key steps in BSP methylation detection?
A: 1) Bisulfite modification of genomic DNA: The efficiency of conversion directly affects the experimental results. Therefore, it is necessary to continuously explore suitable experimental conditions to ensure a high conversion efficiency.
BSP primer design: We design one to two pairs of primers for each target region, and the primer positions should avoid CG sites in the DNA as much as possible. Nested primers may be designed if necessary to amplify the target fragment.
PCR amplification: For target fragments that are difficult to amplify, PCR reaction conditions may need to be adjusted.
Q: Why is an 800 bp target fragment divided into two segments for BSP methylation research?
A: Genomic DNA undergoes fragmentation after bisulfite modification, and the average fragment size is around 500 bp. Therefore, the size of each amplification fragment should not exceed 500 bp.
Q: Can PCR products obtained from BSP methylation amplification be directly sequenced, or is it necessary to construct TA clones for sequencing?
A: Each CG site in genomic DNA can have a different degree of methylation. If C is unmethylated, it is modified to U, while methylated C remains unchanged. If PCR products are directly sequenced, it may result in double peaks at the original C sites, making it difficult to determine the degree of methylation. Therefore, it is necessary to recover the PCR products and perform TA cloning, followed by random selection of 5-10 individual clones for sequencing to calculate the methylation degree at each CG site.
Q: How to analyze the results of BSP methylation sequencing?
A: We use specialized methylation analysis software to analyze the sequencing results of each TA clone. During the analysis, the following information can be obtained:
Similarity between the sequencing sequence and the target sequence.
C-T conversion efficiency.
Confirmation of methylation at each CG site in the target fragment through a black-and-white circle plot.
Determination of the proportion of CG sites that undergo methylation.
Q: How to determine the target region of a gene for methylation analysis using BSP?
A: In BSP methylation detection, it is necessary to determine the region for methylation analysis. Since methylation is closely related to gene regulation, we can refer to relevant literature to analyze the methylation of genes with known transcription regulatory regions. If the position of the transcription regulatory region is unknown, the transcription start site and translation start codon position of the gene can be determined through genomic sequence analysis, and CpG islands can be analyzed. The CpG island near the transcription start site is the preferred region for DNA methylation analysis, typically located in the promoter or 5' UTR region. It should be noted that predictions only identify regions with a high possibility of methylation, and we cannot guarantee that the predicted regions will definitely be methylated.
Q: Why are reported methylation sequences not within CpG islands in some cases?
A: CG dinucleotides are the main sites of methylation, and regions with a high frequency of CG dinucleotides are even more likely to undergo methylation. However, the absence of obvious CpG islands does not mean the absence of methylation. Methylation can also occur in CG regions that are not densely populated with CG dinucleotides.
References
- Li LC. Designing PCR primer for DNA methylation mapping. Methods Mol Biol 2007, 402:371-84.
- Zhang Y, Rohde C,Tierling S, et al.DNA methylation analysis by bisulfite conversion,cloning, and sequencing of individual clones. Methods Mol Biol. 2009,507:177-87.
