We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy
Targeted Metagenomics NGS Analysis
With years of experience in microbial data analysis, CD Genomics provides you with comprehensive targeted metagenomics next-generation sequencing (mNGS) data analysis services and provides new insights for your infectious disease research.
Introduction
In clinical testing laboratories, the scope of routine detection of pathogens in clinical samples includes identification of microorganisms in culture, such as by biochemical phenotyping or matrix-assisted laser desorption/ionization (MALDI) time-of-flight mass spectrometry, and detection of organism-specific biomarkers. For example, use latex agglutination test or enzyme-linked immunosorbent assay (ELISA) antibody test for antigen, or use PCR to perform nucleic acid test on a single sample. However, these detection methods can only detect one or a limited number of pathogens at a time. Targeted detection methods for next-generation metagenomic sequencing are beneficial to increase the number and proportion of pathogen reads in sequence data, and can improve the detection sensitivity of target microorganisms. Targeted NGS analysis methods include: 1. Use highly conservative primers to perform general PCR amplification and detect all microorganisms corresponding to a specific type in clinical samples, such as gene amplification of bacterial 18S rRNA and 16S rRNA. 2. Design primers across the genome to facilitate PCR amplification and amplification, thereby directly recovering the viral genome from clinical samples.
Targeted Metagenomics Next Generation Sequencing Pipeline
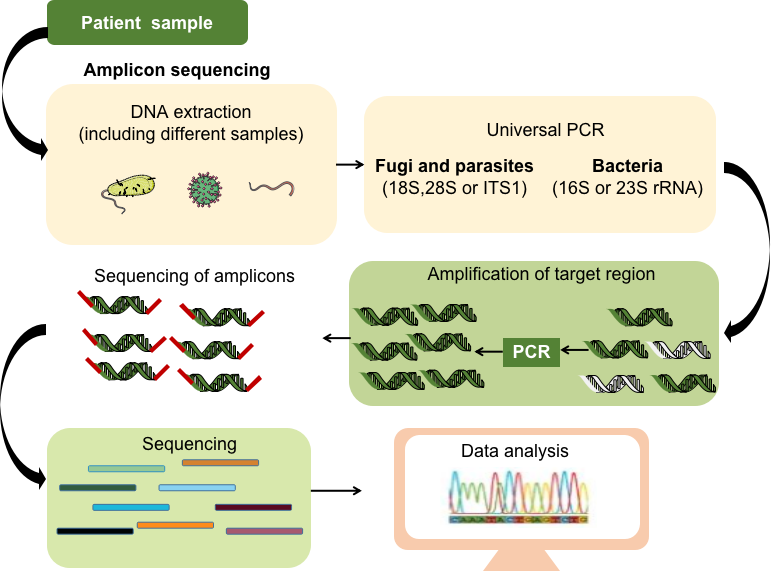 Fig 1. Targeted meta-genomics next-generation sequencing process.
Fig 1. Targeted meta-genomics next-generation sequencing process.
Targeted Metagenomic NGS Analysis Application Areas
- Identify the types of pathogenic microorganisms and assist in the diagnosis of clinical diseases.
- Monitor the changes in the virus, and track the evolution and spread of infectious diseases.
- With the analysis of the diversity of pathogenic microorganisms in different samples, the clinical sample data is enriched to characterize the analysis of antibiotic resistance.
What We Offer
As one of the experienced bioinformatics data analysis providers, CD Genomics provides established, cost-efficient and rapid turnaround analysis services for targeted metagenomics next-generation sequencing data analysis, aiming to help researchers explore the pathogenic microorganisms in infectious diseases or other types of diseases. We provide our clients with the following data analysis services:
- Data analysts evaluate and filter the data.
- Formulate the optimal targeted metagenomics NGS analysis plan.
- Perform data analysis, including develop a personalized analysis plan, or develop a personalized chart.
- Report comprehensive procedures and results.
- Fast project analysis cycle.
Data Ready
Before data analysis, the first thing is to get your data ready. We are able to receive various formats of data for analysis such as raw FastQ/Fasta files, or aligned BAM/SAM files and other intermediate data formats. For targeted metagenomics next-generation sequencing data analysis, in addition to data analysis services, we can also provide customers with corresponding targeted genome high-throughput sequencing and downloading data from public databases. The original data can be from any of the following channels:

For targeted meta-genomics data analysis services, if you have any questions about the data analysis cycle, analysis content and price, please click online inquiry.
What's More
CD Genomics provides customers with one-stop data analysis services, and what customers need to do is to provide raw data and analysis requirements, CD Genomics will develop corresponding analysis strategies based on customer data and provide high-quality reports. In addition, with many years of experience in sequencing services, we can also provide customers with corresponding sequencing services. In addition to targeted metagenomics NGS analysis, we also provide non-targeted metagenomics NGS analysis services. If you have any questions about our targeted metagenomics NGS data analysis service, please feel free to contact us for details.
References
- Gu W, et al. Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection[J]. Annu Rev Pathol.2019 Jan 24; 14:319-338.
- Han D, et al. mNGS in clinical microbiology laboratories: on the road to maturity[J]. Crit Rev Microbiol. 2019 Sep-Nov;45(5-6):668-685.
* For research use only. Not for use in clinical diagnosis or treatment of humans or animals.
Online Inquiry
Please submit a detailed description of your project. Our industry-leading scientists will review the information provided as soon as possible. You can also send emails directly to info@cd-genomics.com for inquiries.
