We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy
Single Cell Regulatory Network Analysis
Introduction
The basis of cell heterogeneity in tissues is the difference in cell transcriptional state, and the specificity of the transcriptional state is determined and maintained stable by gene regulatory networks (GRNs) dominated by transcription factors. Therefore, the analysis of single-cell GRNs can help to dig deeper into the biological significance behind cell heterogeneity, and provide valuable clues for the diagnosis, treatment, and development and differentiation of diseases. However, single-cell transcriptome data has the characteristics of high background noise, low gene detection rate and sparse expression matrix, which brings challenges to traditional statistics and bioinformatics methods to infer high-quality GRNs. With the development of bioinformatics, many software has been developed for the analysis of single-cell regulatory networks. For example, the software SCENIC (Single-cell regulatory network inference and clustering) uses single-cell transcriptome data to analyze regulators, construct gene regulatory networks, and identify stable cell states.
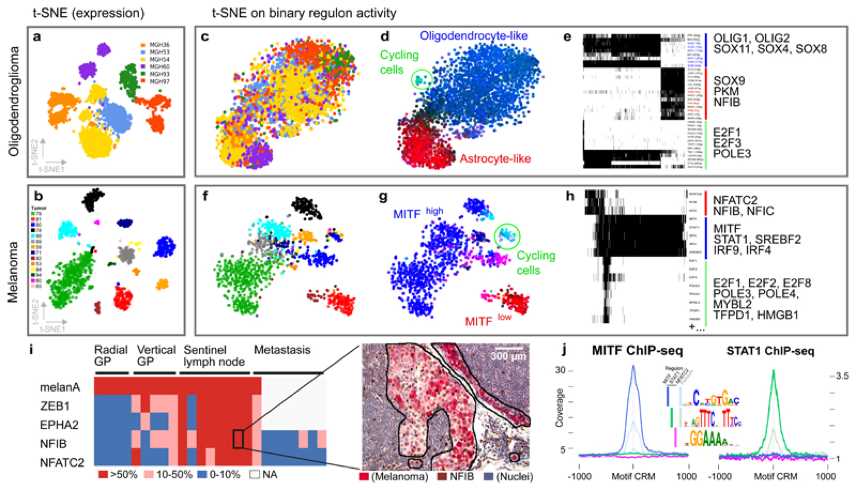 Figure1. Use single-cell RNA-seq data to carry out relevant cell states and gene regulatory networks (GRNs) in cancer. (Aibar S, et al. 2017)
Figure1. Use single-cell RNA-seq data to carry out relevant cell states and gene regulatory networks (GRNs) in cancer. (Aibar S, et al. 2017)
Application Fields
With the maturity of single-cell sequencing technology, single-cell regulatory network analysis has been used in various aspects of the biomedical field:
- Basic research: Such as co-expression module analysis, regulatory element analysis, cell network activity analysis, identification of stable cell states and transcription factors regulation.
- Disease research: Provide valuable clues for biomarker identification, disease diagnosis, personalized treatment, and developmental differentiation research.
Advantages of Single Cell Regulatory Network Analysis
- Removal of batch effects, such as removing batch effects of different batches of clinical samples in different periods, and increasing data analysis sources.
- Analyze complex cell populations.
- Identify cell status.
- Discover potential drivers.
- Study cross-species biological issues.
Data Analysis Technical Route
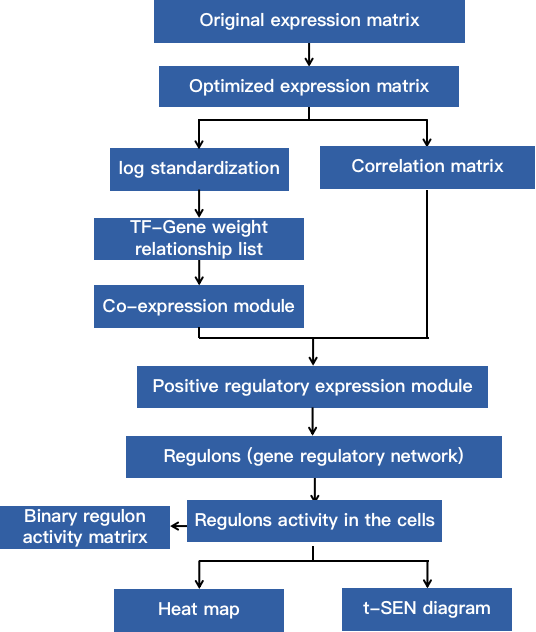 Fig 2. Single cell regulatory network analysis process.
Fig 2. Single cell regulatory network analysis process.
What We Offer
As one of the providers of single cell regulatory network analysis, CD Genomics uses cutting-edge bioinformatics analysis technology and software (such as SCENIC, SCNS, SCODE, SingCellNet, BoolTraineR, and Global GRNs, etc.) to analyze the single cell regulatory network. The raw input data can be downloaded from public databases or produced from a range of sequencing platforms. In addition, we are able to receive various formats of data for analysis such as raw FastQ/Fasta files, or aligned BAM/SAM files and other intermediate data formats. We provide our clients with the following data analysis services:
- Data analysts evaluate and filter the data.
- Formulate the optimal analysis plan.
- Perform data analysis, including develop a personalized analysis plan or personalized charts to summarize results.
- Report complete results.
- Fast project analysis cycle.
Data Ready
Before single cell regulatory network analysis, the first thing is to get your data ready. The raw data or intermediate data can be obtained from the following channels:

If you don't have the data for single cell regulatory network analysis, we can provide relevant help. With many years of sequencing experience, CD Genomics provides customers with different types of single cell sequencing services and download data services. If you have any questions about the sequencing services, data analysis content, turnaround time and price, please click online inquiry.
What's More
Biomedical-Bioinformatics, a division of CD Genomics, provides single cell regulatory network analysis services according to customers' requirements. In addition, we also provide services to obtain raw data from public databases. CD Genomics is happy to work with you at every stage of the research to ensure the best results for your research. For single cell regulatory network analysis, if you have any questions, please feel free to contact us. We have a professional technical support team to provide you the best services. We look forward to working with you.
References
- Aibar S,et al. SCENIC: single-cell regulatory network inference and clustering[J]. Nat Methods. 2017;14(11):1083-1086.
- Sande B, et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis[J]. Nature Protocols. 2020 Jul;15(7):2247-2276.
* For research use only. Not for use in clinical diagnosis or treatment of humans or animals.
Online Inquiry
Please submit a detailed description of your project. Our industry-leading scientists will review the information provided as soon as possible. You can also send emails directly to info@cd-genomics.com for inquiries.
