We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy
Biomarker and Target Discovery
For disease research, a biomarker generally refers to a certain characteristic biochemical index in a general physiological, pathological, or therapeutic process that can be objectively measured and evaluated. By measuring it, the current biological process of the body can be known. With the development of high-throughput sequencing technology, new biomarkers can be used not only as indicators for disease diagnosis and later judgment, but also as therapeutic targets. Biomarkers play an important role in disease research: whether it is to clarify new biomarkers for improved diagnostic methods and drug development, or to verify existing markers in different types and stages of tumors, or to select the best target point transformed into preclinical research, and tumor markers have a great influence on the progress and practical application of tumor research.
Significance of Biomarker Data Analysis
The discovery of a disease-specific biomarker through various experiments and data analysis may be helpful for disease identification, early diagnosis and prevention, and monitoring during treatment. The search and discovery of valuable biomarkers have become an important focus of current research. The analysis of biomarkers by means of bioinformatics can provide data support for experiments to a large extent, thereby speeding up research progress and reducing research costs. With many years of data analysis experience, CD Genomics provides you with various types of biomarker and target discovery data analysis services. Through joint analysis of different omics data, gene expression analysis and interaction network analysis, it helps to identify drug-related targets in the process of disease occurrence and development.
Explore CD Genomics Biomarker and Target Discovery Solutions
CD Genomics provides customers with different types of biomarker and target discovery data analysis services. We provide different data analyses for different types of input raw data, and we will provide different biomarker identification schemes according to different research needs and original data.
Multi-omics Joint Analysis Solutions
A single-group analysis method can provide information on the biological processes of different life processes or disease groups compared with the normal group. However, these analyses often have limitations. Multi-omics joint analysis methods integrate information from several omics levels, provide more evidence for biological mechanisms, and dig out candidate biomarkers from a deeper level. We can provide high-quality multi-omics joint analysis, including: Transcriptome and Metabolome, Proteome and Metabolome, Microbiome and Metabolome, Transcriptome and Proteome, miRNA and mRNA, DNA Methylation-miRNA-mRNA, Genome and Metabolome, LncRNA and Metabolome, mRNA-Proteome-Metabolome, miRNA-mRNA and Protein, and so on.
Example pathway interaction network based on metabolome and transcriptome datasets
Bartel J, et al. PLoS Genetics (2015)

Gene Expression Analysis
Gene expression analysis is to answer some biological questions by analyzing the gene expression matrix. We provide comprehensive data solutions for gene expression profiling, screening, validation, and biomarker analysis based on different types of gene microarray data and next-generation sequencing data, including different analysis directions:
NGS Data & Microarray
- Heat Map
- Volcano Plot
Gene Function Annotation
- HKEGG Pathway Enrichment Analysis
- GO Enrichment Analysis
- COG Enrichment Analysis
Cluster Analysis
- HK-means Clustering Analysis
- Hierarchical Clustering Analysis
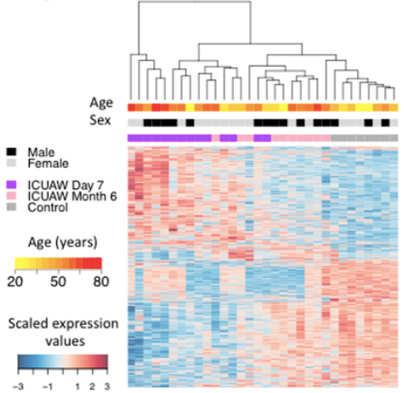
Example heat map based on differentially expressed gene datasets
Walsh C J, et al. Scientific reports (2016)
Biological Network Analysis Solutions
CD Genomics biological network analysis services ensure an in-depth molecular understanding of interaction mode between different small molecules, such as protein, noncoding RNA, transcription factors, metabolites, and so on. To dig the potential biological functions of each target, CD Genomics offers several data analysis approaches:
- PPI Network Analysis
- Residue Interaction Network
- Gene Interaction Network
- CircRNA and Target Gene Interaction Network
- Regulatory Network of Transcription Factors
- Single Cell Regulatory Network Analysis
- Metabolic Network Analysis
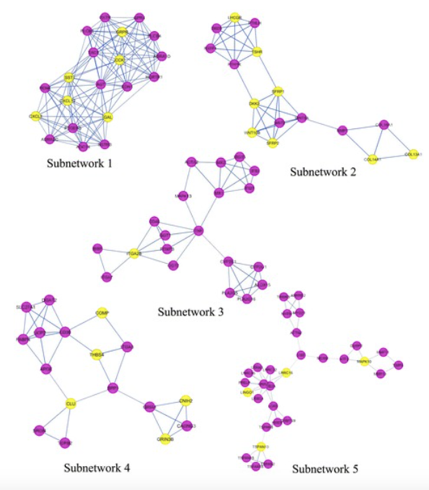
PPI network constructed from DEGs on the basis of human interactome
Kotni M K, et al. Orphanet Journal of Rare Diseases, (2016)
References
- Walsh C J, et al. Transcriptomic analysis reveals abnormal muscle repair and remodeling in survivors of critical illness with sustained weakness[J]. Scientific reports, 2016, 6(1):29334.
- Kotni M K, et al. Gene expression profiles and protein-protein interaction networks in amyotrophic lateral sclerosis patients with C9orf72 mutation[J]. Orphanet Journal of Rare Diseases, 2016, 11(1):148.
- Bartel J, et al. The Human Blood Metabolome-Transcriptome Interface[J]. PLoS Genetics, 2015, 11(6): e1005274.
* For research use only. Not for use in clinical diagnosis or treatment of humans or animals.
Online Inquiry
Please submit a detailed description of your project. Our industry-leading scientists will review the information provided as soon as possible. You can also send emails directly to info@cd-genomics.com for inquiries.
