We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

Navigation
- Home
- Services
- Sequencing Services
- Sanger Sequencing Services
- Agricultural NGS Services
- Animal and Plant Whole Genome Sequencing
- Whole Genome Resequencing
- Reduced-Representation Genome Sequencing (RRGS)
- Targeted Sequencing
- Exome Sequencing
- Animal and Plant De Novo Sequencing
- Chloroplast DNA (cpDNA) Sequencing
- Plant and Animal Mitochondrial Sequencing
- Transcriptome (RNA-seq) Services
- Small RNA Sequencing
- Epigenetic Sequencing
- ChIP-seq Services
- Metagenomics Sequencing Services
- 16S and ITS rRNA Sequencing
- DAP-seq Service for DNA-protein Interaction Research
- Low-Coverage Whole Genome Sequencing (LC-WGS)
- T-DNA Insertion Analysis
- Long-read Sequencing
- SNP Detection
- Plant and Animal Microarray Services
- Gene Mapping in Animals and Plants
- Bioinformatics Analysis Services
- Crop Genome Sequencing
- Livestock Genome Sequencing
- Insect Genome Sequencing
- Sequencing Services
- Products
- Solutions
- Resource
- Company


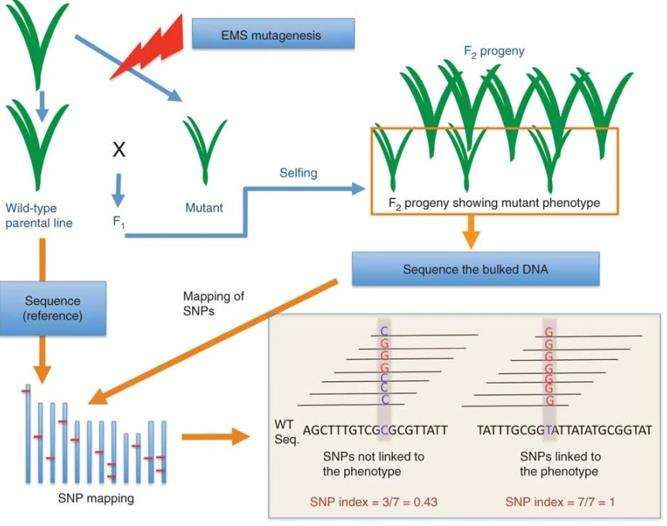 Fig.2. Simplified scheme for application of MutMap. (Abe A, et al., 2012)
Fig.2. Simplified scheme for application of MutMap. (Abe A, et al., 2012)