CD Genomics proprietary GenSeqTM Technology provides Transcriptomic Data Analysis service. We have extensive experience in helping solve a wide variety of bioinformatics problems.
What Is Transcriptomic Data Analysis
Transcriptome data analysis involves processing, interpreting, and analyzing high-throughput sequencing data of RNA transcribed products present in biological organisms. This comprehensive approach yields intricate details about genetic expression patterns active within cells or tissues, encompassing several key stages beyond mere data collection.
Data Pre-processing: This initial stage includes quality control (QC), adapter sequence removal, and alignment to ensure sequencing data quality, laying a reliable foundation for subsequent analyses.
Construction of the Expression Matrix: Following pre-processing, a count matrix enumerates gene reads or quantifies transcripts, forming an expression matrix.
Differential Expression Analysis: Core to transcriptome analysis, this step identifies differentially expressed genes (DEGs) by comparing expression levels between samples, elucidating genes driving phenotypic differences.
Statistical Analysis and Gene Annotation: Utilizing techniques like DESeq2 and edgeR, statistically analyze DEGs and functionally annotate them to understand their biological relevance.
Functional Enrichment Analysis: Conduct enrichment analyses (e.g., Gene Ontology, Pathway) on DEGs to unveil functional characteristics and biological pathways.
Data Visualization and Interpretation: Employ visualization tools (e.g., heat maps, scatter plots) to depict expression patterns, interpreting results against existing biological knowledge to infer biological processes.
Gene Regulation Analysis: Identify and visualize transcription factor families, predict miRNA targets, and analyze miRNA-mRNA-lncRNA regulatory relationships to understand gene regulatory mechanisms.
Transcriptome data analysis demands both bioinformatics expertise and deep biological understanding. Through this process, a comprehensive insight into gene expression patterns and regulatory mechanisms under specific physiological conditions is achieved.
For example, the analysis process of RNA sequencing data is as follows:
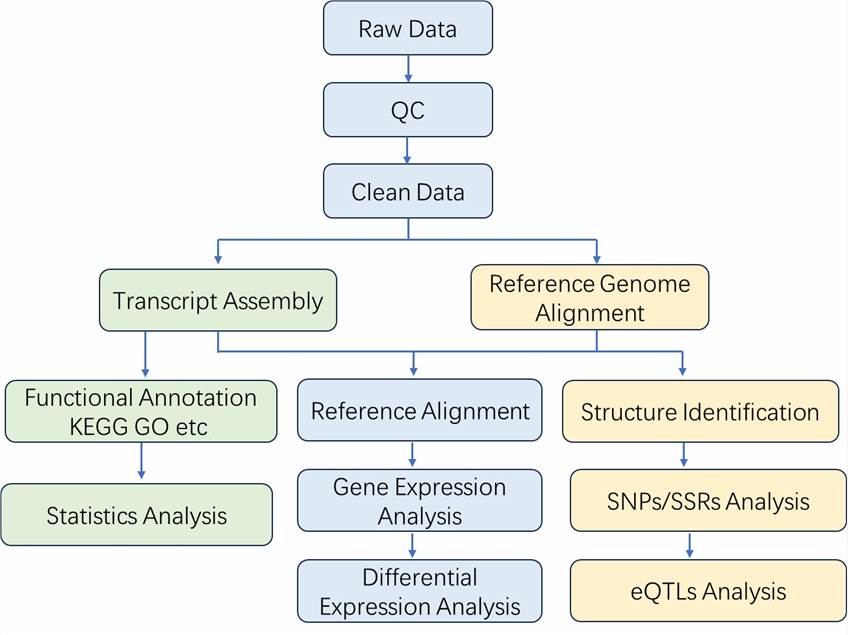
Our Transcriptomic Data Analysis Service
The transcriptome is the complete set of transcripts in a cell, tissue, or whole organism, for a specific developmental stage or physiological condition. Transcriptomic Data Analysis involves characterization of all transcriptional activity (coding and non-coding), or a select subset of RNA transcripts within a given sample. The analysis of transcriptomes allows the identification of candidate genes and expressed markers associated with traits of interest. We offer the following transcriptomic data analysis services:
-
Next-Generation Sequencing Transcriptomics Data Analysis
-
Next-generation sequencing (NGS) transcriptomics represents a robust approach for exhaustively dissecting the transcriptome. This cutting-edge technology furnishes unparalleled granularity and insight into studying gene expression and regulatory networks, finding extensive applications within diverse scientific domains encompassing biomedical research, agriculture, environmental science, and beyond. CD Genomics offers next-generation transcriptomics sequencing data analysis, which primarily includes RNA-Seq Data Analysis, Small RNA Sequencing Data Analysis, LncRNA Sequencing Data Analysis, Ribo-seq Data Analysis and etc.
With our Next-Generation Sequencing Transcriptomics Data Analysis service, we are able to provide:
Raw data QC
Alignment and TPM/RPKM/FPKM-based quantitation
Expression analysis
Statistics of SNPs/Indels
Alternative splicing analysis
GO and KEGG annotation
Different sequencing services offer corresponding data analysis options. You can explore the transcriptomics sequencing services of interest to find more detailed information.
-
-
Alternative splicing produces diverse protein isoforms. PacBio Iso-Seq captures full-length transcripts, aiding in identifying isoforms and novel transcriptional overlaps. This technology complements short-read RNA-Seq, enabling comprehensive gene expression analysis and network studies.
With our Full-Length Transcripts Sequencing Data Analysis service, we are able to provide:
Transcriptome with no Reference:
Full-length Isoform analysis, including Full-length Isoform correction, Classification, Reduced Redundancy
Transcriptome annotation, including Gene Ontology, KEGG pathway, KOG or COG, Swissport
Gene Structures analysis, including alternative splicing, LncRNA, SSR, CDS
Map to Reference
Gene expression level analysis
Differentially expressed gene analysis
Differentially expressed transcriptome KEGG enrichment analysis
Differentially expressed gene heatmap
… (more upon request)
Transcriptome with Reference:
Full-length Isoform analysis, including Reference Genome mapping, Full-length Isoform correction, Classification, Reduced Redundancy
Transcriptome annotation, including Gene Ontology, KEGG pathway, KOG or COG, Swissport
Gene Structures analysis, including alternative splicing, LncRNA, SSR, CDS, novel transcriptome prediction, Identification of fusion gene
… (more upon request)
-
-
Third-generation sequencing, exemplified by Oxford Nanopore's MinION device, produces long reads (1–100kb) by threading single-stranded DNA through nanopores. This real-time sequencing method offers continuous sequencing with minimal reagent consumption, overcoming short-read assembly challenges and revolutionizing genomics research.
With our Nanopore Full-Length Transcripts Sequencing Data Analysis service, we are able to provide:
Oxford Nanopore Technologies Long Read Processing
Remove redundant
Find fusion transcript
Structure analysis
Transcription factors prediction
lncRNA analysis
Gene functional annotation
SNP calling
Quantification of gene/transcript expression levels and Differential expression analysis
Functional enrichment analysis
PPI (Protein Protein Interaction)
-
-
10x Spatial Transcriptome Sequencing enables simultaneous analysis of gene expression and spatial distribution within tissues or cells, offering insights into cellular organization and interactions.
With our 10x Spatial Transcriptome Sequencing Data Analysis service, we are able to provide:
Raw sequence data
Sequencing data quality assessment and filtering
Spot quality control
Data alignment
Data standardization
Spot clustering
Spot subpopulation analysis
Marker analysis
Cell type identification
Anatomical region annotation
Cell communication analysis
Inter-sample differential analysis
Inter-annotation region differential analysis
Advantages of Transcriptomic Data Analysis
- Transcriptomic data analysis, a potent investigative tool, delivers holistic insights into gene expression patterns within biological systems, thereby empowering researchers with a profound comprehension of intricate regulatory mechanisms and fresh revelations concerning cellular processes.
- Professionals well-versed in the realm of transcriptomic data analysis showcase prowess across diverse analytical realms such as experimental design, data preprocessing, differential expression analysis, and functional interpretation. Their acumen guarantees accurate and insightful interpretations of transcriptomic data, thereby underpinning the foundation for robust conclusions and groundbreaking discoveries.
- The services extended within the realm of transcriptomic data analysis offer rapid, precise, and cost-effective solutions for gene expression data analysis. Consequently, researchers can swiftly access top-tier transcriptomic data without any compromise on turnaround time or budgetary constraints.
- The adaptability of transcriptomic data analysis platforms equips researchers to navigate through an array of datasets stemming from varied experimental conditions, sample types, and species. Such flexibility empowers researchers to engage with a wide spectrum of research questions and tailor their analyses to suit specific experimental demands.
- The progress of biological research is propelled by the insights gleaned from transcriptomic data analysis, unlocking novel understandings of gene expression regulation and cellular functions. This analytical methodology plays a pivotal role in advancing our knowledge of disease mechanisms, facilitating drug discovery, and fostering the evolution of personalized medicine modalities.
- The services encompassed within transcriptomic data analysis showcase a customizable analytical framework that's tailored to the distinct requisites of each research undertaking. Researchers are provided with the autonomy to select specific analysis methods, parameters, and criteria, thus aligning with their research goals and hypotheses.
- Practitioners in the field of transcriptomic data analysis embrace a consultative approach, engaging in close collaboration with researchers to delineate optimal analysis strategies. Our expert counsel aids researchers in navigating complex analytical workflows and ensuring precise interpretation of results.
Application of Transcriptomic Data Analysis
- Gene Expression Analysis: Transcriptomic data analysis enables the precise identification and quantification of gene expression levels, unveiling distinctive gene expression patterns in diverse biological contexts.
- Biological Pathway Investigation: By leveraging transcriptomic data, researchers can reconstruct metabolic pathways, signaling cascades, and intricate cellular networks to decipher the mechanisms governing gene regulation.
- Co-expression Gene Clustering: Through advanced clustering analyses, co-expressed genes can be discerned, contributing to the understanding of genome-wide gene co-regulation patterns during biological processes.
- Functional Gene Inference: The integration of transcriptomic data and bioinformatics tools facilitates the prediction of gene functions in cellular processes, metabolic pathways, and disease progression.
- Discovery of Novel Genetic Elements: Exploration of uncharted genes or regulatory RNAs through transcriptomic data mining may reveal previously unknown biological processes or potential therapeutic targets.
- Environmental Adaptation Mechanisms: Analysis of transcriptomic data delves into gene regulatory networks orchestrating biological adaptation to varying environmental conditions, elucidating organismal responses in distinct environments.
- Drug Response Assessment: By scrutinizing gene expression profiles post-drug intervention through transcriptomic data, the underlying mechanisms of drugs, potential resistance patterns, and individualized variations in drug responses can be unveiled.
- Disease Biomarker Identification: Comparing gene expression profiles between patients with specific diseases and healthy cohorts using transcriptomic data aids in pinpointing potential biomarkers for early disease detection and therapeutic intervention.
Analysis Cases
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines


