Available on both Illumina and PacBio Platforms, CD Genomics offers the accurate and affordable mitochondrial DNA sequencing solution you're looking for.
Introduction of mtDNA Sequencing
Mitochondrial DNA (mtDNA) is a compact, double-stranded circular genome of 16,569 bp with a cytosine-rich light (L) chain and a guanine-rich heavy (H) chain. Mitochondria play a very important role in important cellular functions. In addition to producing over 90% of the energy required by a cell, mitochondria also generate reactive oxygen species (ROS) and participate in apoptosis and other important cellular functions. The mutant mtDNA and the wild type may co-exist as heteroplasmy, and cause human disease, including cancer, heart disease, diabetes, Alzheimer's disease, Parkinson's disease and hypertension. Because there is considerable clinical variability between mitochondrial disorders and many patients who exhibit phenotypes that overlap diseases, often diagnosis may only be confirmed by identification of a pathogenic mtDNA variant through molecular genetic testing of DNA extracted from a blood sample. Mitochondrial DNA sequencing is a useful tool for researchers to study human diseases, and be also used in population genetics and biodiversity assessments.
Mitochondrial DNA sequencing is available on both Illumina's MiSeq, and PacBio Sequel System. Illumina next-generation sequencing (NGS) has the potential to transform mtDNA analysis. Take advantage of PacBio's long reads, CD Genomics present amplification-free, full-length sequencing of linearized mtDNA. Full-length sequencing allows variant phasing along the entire mitochondrial genome, identification of heteroplasmic variants, and detection of epigenetic modifications that are lost in amplicon-based methods.
Advantages of Our mtDNA Sequencing Service
- Capture efficiency & high coverage, 100% amplicon coverage of all regions of the mitochondrial genome
- Highest accurate detection of rare variants in mtDNA
- Novel bioinformatics analysis programs and pipelines
- Well-experienced personnel
Application of mtDNA Sequencing
- Analysis of mitochondrial-related diseases
- Species evolution research
- Systematics and evolutionary biology research
- Population genetics and conservation biology research
- Human genetics research
- Environmental exposure and toxicology research
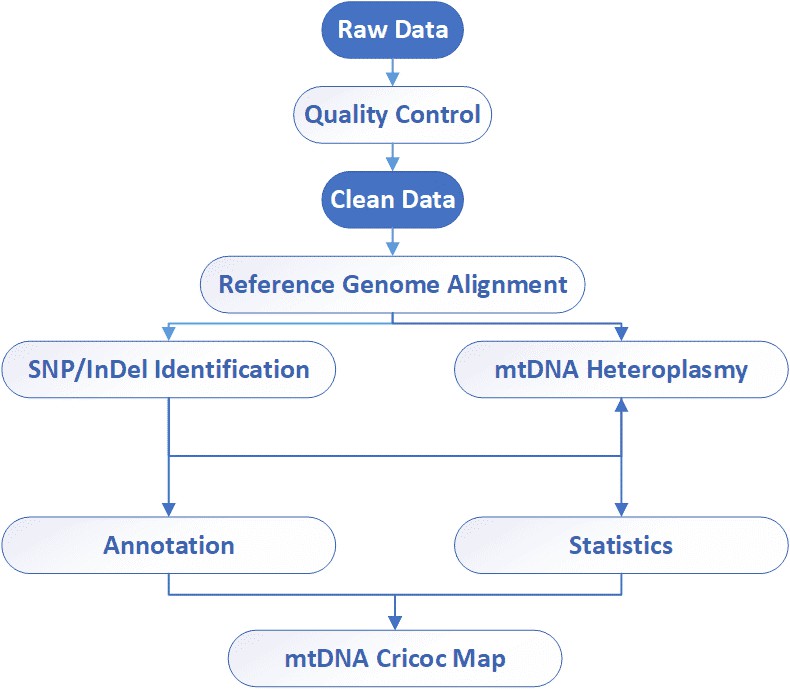
mtDNA Sequencing Workflow

Service Specifications
Sample requirements
|
|
Sequencing
|
|
Data Analysis
|
Analysis Pipeline
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in mtDNA Sequencing for your writing (customization)
CD Genomics proffers cost-efficient mitochondrial DNA sequencing strategies. These encompass the preparation, sequencing, and in-depth analysis of the complete mitochondrial DNA (mtDNA) genomic sequences derived from pristine and intact DNA samples. Our expertise enables the achievement of exceptional sensitivity, specificity, and accuracy in the identification of mtDNA mutations and evaluation of heteroplasmy levels. For additional information and a comprehensive quotation, please do not hesitate to contact us.
Reference:
- Yao Y, Nishimura M, Murayama K, et al. A simple method for sequencing the whole human mitochondrial genome directly from samples and its application to genetic testing. Scientific Reports, 2019, 9(1): 17411.
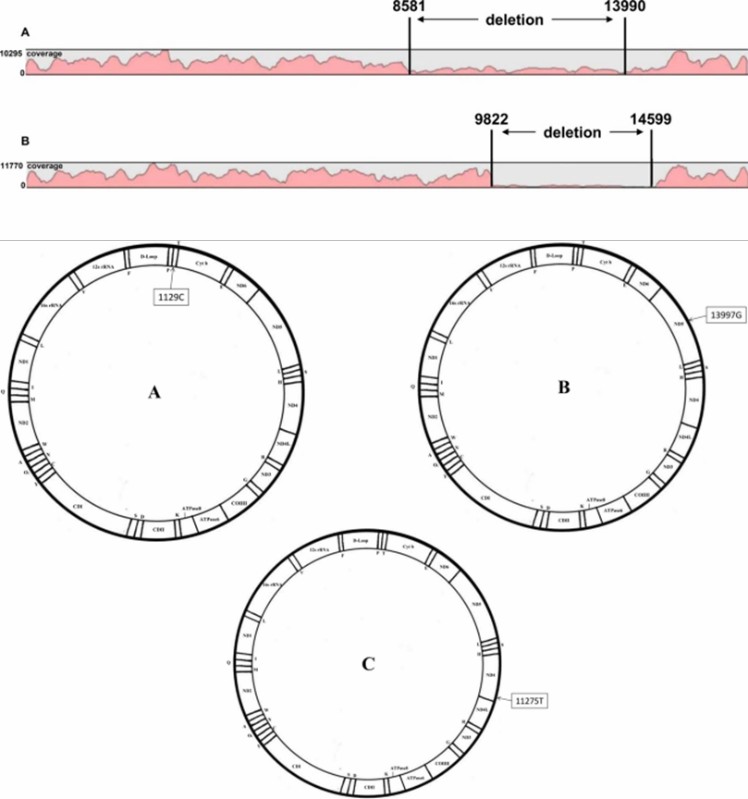 Mapping and coverage results of mtDNA sequencing, as well as the display of mtDNA pathological profiles. (Yao et al., 2019)
Mapping and coverage results of mtDNA sequencing, as well as the display of mtDNA pathological profiles. (Yao et al., 2019)
1. Why are mtDNA sequences used rather than autosomal DNA sequences?
The mitochondrial genome presents a plethora of distinctive features, including high variabilities, swift evolution, and a complete lack of recombination. The mitochondrial genome is relatively diminutive, and the mutation rate of mitochondrial DNA usually exceeds that of chromosomal DNA throughout evolution. In multicellular organisms, each cell typically contains multiple mitochondria, usually outnumbering the cell nuclei. Consequently, compared to nuclear DNA, mitochondrial DNA extraction from samples is considerably more feasible, providing an amplifiable amount of DNA for further analysis. Furthermore, the mitochondrial genome plays a pivotal role in certain hereditary diseases.
2. Does whole genome sequencing include mitochondrial DNA?
Whole-genome sequencing typically encompasses mitochondrial DNA (mtDNA). Despite notable differences from nuclear DNA, such as its smaller size and lack of recombination, mtDNA is commonly considered as part of whole-genome sequencing. Researchers employ specific sequencing methods and technologies during whole-genome sequencing to ensure the integrity and coverage of mtDNA.
3. What are the challenges associated with mtDNA sequencing
Firstly, the abundance of mitochondrial DNA within cells can potentially lead to contamination issues. This is further compounded by the presence of heteroplasmy, where multiple variants of mitochondrial DNA coexist within a single individual, adding complexity to data analysis. In addition, the rapid evolution of mitochondrial DNA can render sequence alignments challenging across different species.
4. Is the sample for human mitochondrial capture sequencing DNA or mtDNA?
The sequencing technique of human mitochondrial capture employs probes specifically targeting mitochondria, thereby obviating the need for isolating mtDNA. Consequently, the process can be facilitated merely with the provision of blood or DNA samples.
Genetic Control over mtDNA and Its Relationship to Major Depressive Disorder
Journal: Current Biology
Impact factor: 9.494
Published: December 21 2015
Backgrounds
Previous studies have indicated that the quantity of mitochondrial DNA (mtDNA) experiences alterations in response to external stressors, suggesting a potential correlation with stress-associated disorders such as Major Depressive Disorder (MDD). To explore the genetic regulation mechanisms of mtDNA and its connection with MDD, we conducted an analysis utilizing the low-coverage whole-genome sequencing data of 10,442 Chinese Han female MDD patients. This was performed in order to carry out a genome-wide association study (GWAS) on mtDNA levels.
Methods
- Saliva samples
- DNA extraction
- Mitochondrial DNA (mtDNA) Sequencing
- Whole-genome sequencing
- Illumina Hiseq
- Estimation of mtDNA Copy Number
- Linkage Disequilibrium Adjusted Kinship
- GWAS Using a Linear Mixed Model
- Estimation of Genetic Correlation between Amount of mtDNA and MDD
- Homoplasmic and Heteroplasmic Variants in the mtDNA
Results
This study utilizes low-coverage sequence data from 10,442 Chinese women to compute the standardised read count mapped to the mitochondrial genome as a proxy for mtDNA quantity. Two loci contributing to mtDNA levels were identified: one in the TFAM gene on chromosome 10, and the other on the CDK6 gene on chromosome 7. These gene loci replicate within an independent array. CDK6 emerges as a novel molecule involved in mtDNA control. The incidence of heteroplasmy is increased in women with Major Depressive Disorder (MDD), and this escalation may be induced by stress, as demonstrated using a mouse experimental paradigm. Furthermore, at least one heteroplasmic variant significantly correlates with fluctuations in mtDNA quantity. These findings suggest both copy count and sequences of mitochondrial genome play vital roles in an organism's stress response.
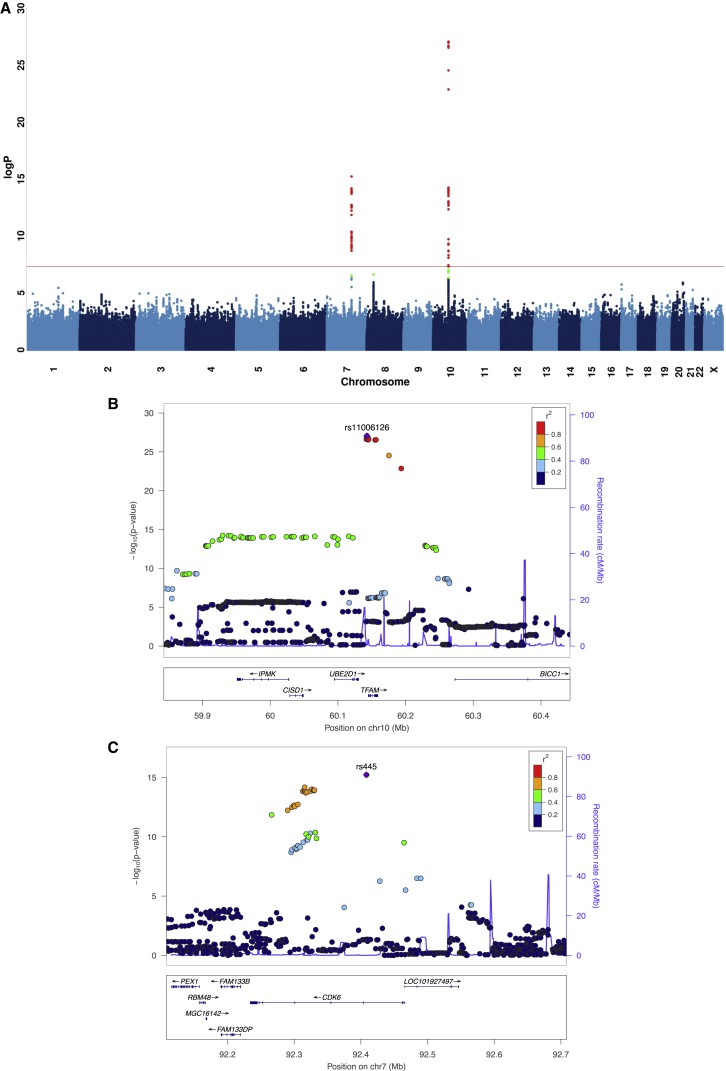 Figure 1. Two Loci Associated with mtDNA.
Figure 1. Two Loci Associated with mtDNA.
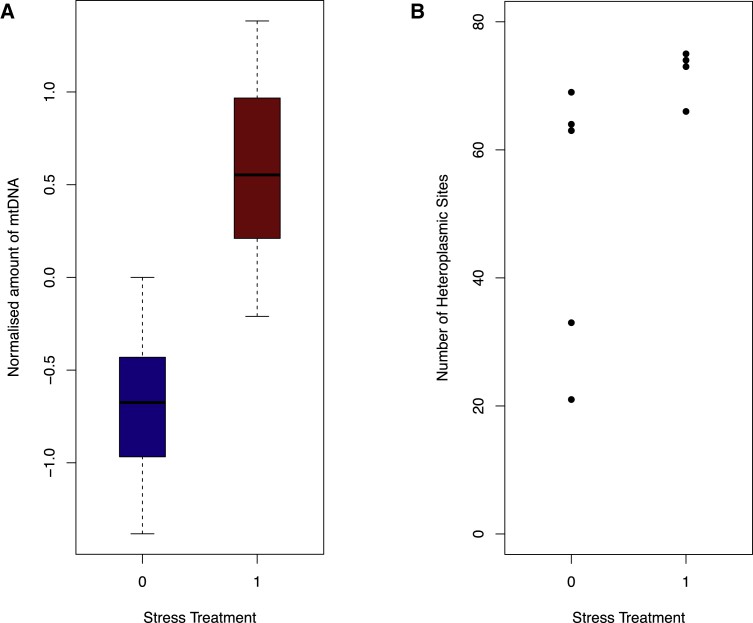 Figure 2. Heteroplasmy Counts in Stressed and Control Mice.
Figure 2. Heteroplasmy Counts in Stressed and Control Mice.
Conclusion
The findings from this research illustrate that loci near TFAM and CDK6 can instigate variations in mtDNA quantities. Notably, in severe cases of depression, mutations have been observed to accumulate within mitochondrial DNA. Animal model experiments have demonstrated the induction of heterogeneity due to chronic stress. Significantly, the amount of mtDNA is associated with loci-specific heterogeneity.
Reference:
- Cai N, Li Y, Chang S, et al. Genetic control over mtDNA and its relationship to major depressive disorder. Current Biology, 2015, 25(24): 3170-3177.


 Sample Submission Guidelines
Sample Submission Guidelines