CD Genomics proprietary GenSeqTM Technology provides Genomic Data Analysis service. We have extensive experience in helping solve a wide variety of bioinformatics problems.
What Is Genome Data Analysis
Genome data analysis encompasses the intricate process of scrutinizing and interpreting the vast repository of genetic information nestled within an organism's genome. The genome, comprising the entirety of DNA, spanning both coding and non-coding regions, constitutes an individual's genetic blueprint. To unravel the hidden insights embedded in genomic data, sophisticated bioinformatics tools and computational techniques are harnessed, facilitating the extraction of meaningful knowledge.
The fundamental objective of genome data analysis revolves around delving into diverse aspects of the genome, including the identification of genes, regulatory elements, and functional components. Moreover, it entails comprehending genetic variations and exploring genetic relationships within populations and individuals. The significance of genome data analysis extends across a wide array of research disciplines, encompassing genomics, genetics, personalized medicine, evolutionary biology, and biotechnology.
Numerous essential tasks constitute the landscape of genome data analysis:
Sequence Alignment: A pivotal process that involves comparing and aligning DNA sequences with reference genomes to reveal both similarities and dissimilarities.
- Variant Calling: The discernment and annotation of genetic variations, such as single nucleotide polymorphisms (SNPs) and insertions/deletions (indels).
- Gene Expression Analysis: Unraveling the intricacies of gene expression patterns across different tissues or under varying conditions.
- Functional Annotation: Illuminating the roles and functions of genes and regulatory elements to unravel their biological significance.
- Comparative Genomics: Uniting genomes of distinct species in a comparative framework to study evolutionary relationships and identify conserved regions.
- Epigenetic Analysis: Investigating DNA and histone modifications to comprehend gene regulation and decipher epigenetic changes.
- Metagenomics: Exploring genetic material from intricate microbial communities to discern and characterize diverse species.
- Pathway Analysis: Scrutinizing interactions between genes and proteins to gain insight into biological pathways and intricate networks.
Altogether, genome data analysis assumes an indispensible role in decoding the genetic information inscribed within an organism's DNA. Its significance extends far and wide, invigorating the advancement of our knowledge in genetics and biology. Moreover, it finds practical applications across diverse domains, including medical diagnostics, drug development, and agricultural research.
Workflow of Genomic Data Analysis
Genomic data analysis embodies an essential step in biological research, involving the processing and interpretation of extensive data derived from biological specimens. Irrespective of the precise mode of analysis, there prevails a universal pattern to data analysis. Commonly, the procedure encompasses the collection of data, quality assessment and cleaning, processing, modeling, visualization and reporting.
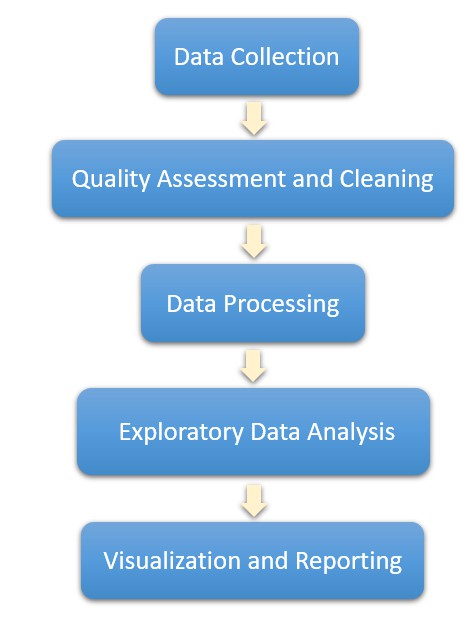
Data Collection: The acquisition of data encompasses a meticulous gathering process from diverse origins, including high-throughput sequencing experiments. This entails the sequencing of DNA or RNA samples to procure genomic data. Additionally, the utilization of publicly accessible datasets and specialized databases enriches the comprehensive compilation of information.
Data Quality Check and Cleaning: Rigorous quality assessments are conducted to meticulously identify and rectify any anomalies inherent within the dataset. This meticulous scrutiny involves the detection and mitigation of missing values, noise, or technical biases. Specifically within genomic data, efforts are directed towards discerning and eliminating low-quality bases, thereby enhancing the precision of read mapping procedures.
Data Preprocessing: At this juncture, the primary endeavor involves the meticulous curation and organization of raw data, priming it for subsequent analytical scrutiny. This process encompasses the discernment and elimination of low-quality reads, rectification of technical biases, attenuation of noise, and culling superfluous sequence fragments. Furthermore, data preprocessing encompasses the alignment of genomic data, the anchoring of sequencing reads to a reference genome, and the precise quantification of gene or region expression levels.
Exploratory Data Analysis and Modeling: In this phase, we leverage the refined dataset to delve into the intricate interplay between variables and discern disparities among samples. Employing a fusion of statistical and machine learning methodologies, we endeavor to unravel latent patterns and structural nuances embedded within the data matrix. The overarching goal of exploratory data analysis is to unearth fundamental motifs, outliers, and evolving trends, thereby laying the conceptual groundwork for subsequent modeling endeavors. To facilitate comprehension and insight generation, a plethora of visualization tools including histograms, scatter plots, and heatmaps are judiciously employed to illuminate data distributions and interrelationships.
Visualization and Reporting: During this phase, the culmination of analytical efforts transitions into visually comprehensible representations, harnessing a blend of conventional data visualization methodologies and bespoke visualizations crafted for genome data analysis. These visualizations function as conduits, facilitating the communication of intricate patterns and insights unearthed throughout the analytical process.
Our Genomic Data Analysis Service
With sequencing technologies now producing millions of high quality reads per run, working with sequence data has become a significant obstacle for many researchers. At CD Genomics, we have staff of dedicated bioinformaticians with extensive experience in overcoming these and a variety of other challenges that researchers face every day. We offer the following genomic data analysis services:
-
-
De novo sequencing can be used to sequence uncharacterized genomes if there is no available reference sequence or known genomes if significant variations are expected.
The general strategy of de novo sequencing analysis is to align and merge short fragments derived from a much longer DNA sequence in order to reconstruct the original sequence. de novo sequencing projects usually take multiple libraries and multiple rounds of finishing to get a complete genome sequence.
With our de novo Sequencing Data Analysis service, we are able to provide:
Generation of high-quality reference genome assemblies
Structural and functional annotation of genes
Identification and phylogenetic analysis of gene families (i.e. R-genes)
Prediction of biosynthetic
Pathways
-
Annotation and Gene Prediction
-
Once the genome of an organism has been sequenced and assembled, genes must be identified in order to understand the functional content of the genome. For this reason gene prediction and annotation are among the most important steps of a genomic project. The goal of annotation is to identify the key features of the genome, in particular protein-coding genes and their products.
We are able to use data coming from de novo or resequencing projects to perform gene predictions, small and large non coding RNA annotation, identification of specific gene and protein families and pathways.
-
-
Once you have the reference sequence for an organism, you can utilize next-generation sequencing to perform comparative sequencing or resequencing to characterize the genetic variations in individuals of the same species or between related species.
With our Genome Resequencing Data Analysis service, we are able to provide:
Identification of small structural variations (SVs) such as SNPs and DIPs;
Identification of large SVs like deletions, insertions, duplications, inversions, genomic rearrangements, Copy Number Variations (CNV) and Presence Absence Variations (PAV);
Genome consensus reconstruction;
Genome Wide Association Studies (GWAS);
High-throughput genotyping;
Molecular evolution studies.
-
Targeted & Exome Sequencing Data Analysis
-
Due to high sequencing and data management costs associated with whole genome resequencing, Targeted Resequencing provides a time- and cost-effective alternative. Targeted Resequencing, including Exome Sequencing, primarily focuses on detecting SNP and small Indels.
With our Targeted & Exome Sequencing Data Analysis service, we are able to provide:
Identification of small structural variations (SVs) such as SNPs and DIPs;
Identification of large SVs like deletions, insertions, duplications, inversions, genomic rearrangements, Copy Number Variations (CNV) and Presence Absence Variations (PAV);
Genome Wide Association Studies (GWAS);
High-throughput genotyping;
High-throughput development of molecular markers;
Molecular evolution studies.
-
-
Metagenomics is the study of genomes contained within an entire microbial community. Metagenomic sequencing focuses on microbial community diversity analysis, gene composition and function, as well as metabolic pathways associated with the specific environment.
With our Metagenomic Sequencing Data Analysis service, we are able to provide:
Analysis of species composition and abundance
Genome components analysis
Generate non-redundant gene catalog
Gene functional annotations
Comparative analysis among samples
Advantages of Genomic Data Analysis
- Comprehensive Solutions: Genome data analysis presents holistic solutions, affording researchers profound insights into the entirety of an organism's genetic blueprint.
- Expertise Across Sequencing Aspects: Professionals proficient in genome data analysis boast multifaceted expertise spanning various facets of sequencing, encompassing experiment design, target enrichment library construction, and tailored bioinformatics analysis, thereby ensuring meticulous and precise outcomes.
- Fast, Accurate, and Affordable Services: Genome data analysis delivers expeditious, precise, and cost-effective services, facilitating accessibility to top-tier genomic data devoid of compromise on quality or expeditious turnaround.
- Versatility in Sample Handling: With sophisticated capabilities, genome data analysis adeptly manages a diverse array of samples, spanning human, animal, plant, and microbial sources, thereby ensuring adaptability to diverse research exigencies.
- Advancement of Genomics Research: Genome data analysis serves as a catalyst for the progression of genomic inquiry, granting researchers access to state-of-the-art technologies and methodologies, empowering them to achieve breakthrough discoveries.
- Flexible Service Customization: Genome data analysis offers customizable services tailored to individual project specifications, ensuring that researchers receive bespoke analysis solutions meticulously aligned with their unique objectives.
- Consultative Approach for Optimal Solutions: Practitioners of genome data analysis adopt a consultative stance, discerningly identifying optimal, cost-efficient solutions tailored to research imperatives, proffering expert guidance and counsel to realize optimal outcomes within fiscal constraints.
Application of Genomic Data Analysis
- Healthcare and Medical Field: The analysis of genomic data holds immense potential applications in the prevention, diagnostic evaluation, and therapeutic planning for myriad diseases, driving forward the arenas of ancestral tracing and personalized medicine.
- Agricultural Sector: Within the context of agricultural biotechnology, leveraging the power of genomic data analysis can catalyze efforts towards improving crop quality, elevating yield, and enhancing resistance to disease and adaptability to climate change.
- Environmental Protection: Moreover, genomic data analysis has proven to be indispensable in conducting research on environmental microorganisms and for the preservation of threatened species.
- Biotechnology: Advancing the utilization of genomic data analysis facilitates more effective strategizing within the biotechnological realm, notably in genetic engineering, cell engineering, and similar disciplines.
- Forensic Sciences: By conducting genomic data analysis of biological specimens found at crime scenes, individual identification is made possible, thereby providing a powerful tool for criminal investigation efforts.
For Research Use Only. Not for use in diagnostic procedures.


 Sample Submission Guidelines
Sample Submission Guidelines

