As seasoned purveyors of Next-Generation Sequencing (NGS) and third-generation sequencing services, our commitment lies in furnishing unparalleled quantities of sequencing data to underpin swift and pioneering analytical methodologies, all while offering economical resolutions. Profoundly adept in the realm of DNA 6mA sequencing, we harness cutting-edge high-throughput sequencers, meticulous sequencing methodologies, and sophisticated bioinformatics analysis pathways to furnish our clientele with assured and impartial services.
The Introduction of DNA 6mA Sequencing
DNA methylation is implicated as an epigenetic mark in various important processes in eukaryotes. 5-methylcytosine (5mC) is the most predominant DNA methylation modification in eukaryotes and has been acknowledged as the best-characterized epigenetic markers. N6-methyladenine (6mA) was previously believed to exist only in prokaryotes, unicellular eukaryotes, and plants. In recent years, 6mA in DNA has been defined as another important epigenetic and epitranscriptomic marker in higher eukaryotes. A significant decrease in 6mA levels has also been reported in a variety of cancer cells (unpublished data).
CD Genomics uses multiple strategies to validate genome-wide methylation status and abundance of individual 6mA sites. Long read sequencing developed by Pacbio and Nonapore can be used to detect 6mA in different genomes. 6mA DNA immunoprecipitation followed by deep sequencing (DIP-Seq) is another key tool to identify regions in the genome that contain 6mA.
Methods of DNA 6mA detection
In the realm of 6mA detection, several methodologies stand out:
6mA-IPseq: This method involves immunoprecipitation of methylated DNA fragments using 6mA antibodies followed by sequencing. It's cost-effective but lacks precise methylation site localization due to antibody limitations and potential false positives from unmethylated DNA and RNA interference.
6mA-REseq: Based on restriction enzyme digestion of unmethylated motifs, this method assesses methylation status based on the ratio of internal to terminal motifs. It's limited by specific restriction sites and potential false positives due to incomplete digestion.
HPLC-MS/MS: High-performance liquid chromatography coupled with tandem mass spectrometry delivers exceptional sensitivity and specificity in identifying 6mA. This method enables precise quantification of nucleosides; however, it is susceptible to interference from bacterial contamination, necessitating stringent experimental controls.
SMRT: Single-molecule real-time sequencing harnesses DNA polymerases and fluorescence-labeled nucleotides to directly detect 6mA modifications. While offering considerable power, it is susceptible to elevated false positive rates, particularly in scenarios with low 6mA concentrations. Furthermore, it lacks the capability to differentiate between 6mA and 1mA modifications.
Deep Learning Predictive Models: Diverse deep learning models have emerged for the prediction of 6mA sites, For example, DNA6mA-MINT, i6mA-stack, Deep6mA, and others. These models boast impressive accuracy; however, their applicability across species may be limited, necessitating ongoing optimization efforts.
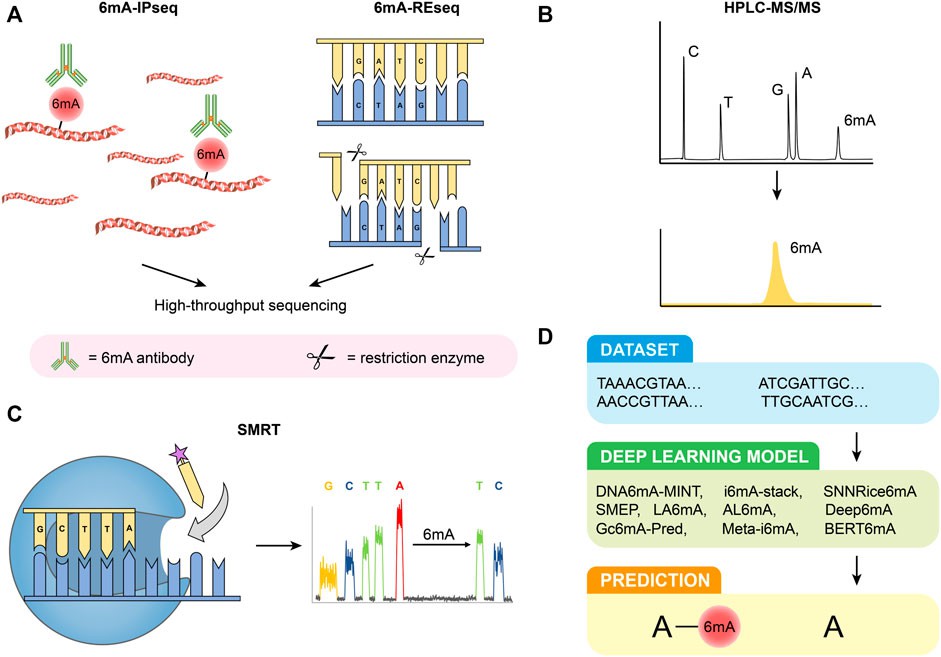 Figure 1. Detection methods of 6 mA. (A) 6 mA-IPseq and 6 mA-REseq. (B) HPLC-MS/MS. (C) SMRT. (D) Deep learning predictive model. (Li et al., 2022)
Figure 1. Detection methods of 6 mA. (A) 6 mA-IPseq and 6 mA-REseq. (B) HPLC-MS/MS. (C) SMRT. (D) Deep learning predictive model. (Li et al., 2022)
Advantages of DNA 6mA Sequencing
- Decoding Genomic DNA Methylation Patterns: 6mA is a crucial modification in genomic DNA, implicated in the regulation of gene expression and genomic stability. Sequencing for 6mA affords insights into its distribution pattern, modification levels, and its association with biological processes, thereby advancing our understanding of its function and significance.
- High-resolution and Whole-genome Coverage: The third-generation sequencing technology enables direct sequencing of individual DNA molecules, providing a higher resolution than traditional second-generation sequencing. Consequently, it allows for the accurate detection of 6mA distribution and offers unrestricted access across the genome, enabling whole-genome 6mA detection.
- High Sensitivity and Specificity: Third-generation sequencing boasts exceptional sensitivity and specificity, enabling precise detection of 6mA modifications while discerning other related modifications. This capability facilitates efficient enrichment and detection of 6mA, thereby fostering a deeper comprehension of its biological function and significance.
- Capturing Long-range Information: The ability of third-generation sequencing to capture larger DNA fragments in a single read enhances our understanding of the long-range interactions and regulatory mechanisms of 6mA at a genomic scale.
- Efficient Enrichment and Detection: The 6mA DIP-Seq technique harnesses the specificity of 6mA antibodies to enrich methylated DNA fragments and probes them using deep sequencing technology. This approach facilitates efficient enrichment and detection of 6mA, enabling a precise determination of its loci and distribution patterns.
- Cost-Effectiveness: Relative to other comparable methods, 6mA DIP-Seq proves to be less costly, positioning it as an economically viable choice, especially for large-scale 6mA sequencing projects or those constrained by budget.
- Ease of Operation: The relatively simplified and straightforward procedure of 6mA DIP-Seq eliminates the need for complex preprocessing steps, making it more accessible to researchers while ensuring reliable 6mA sequencing data within a shorter time frame.
Application of DNA 6mA Sequencing
Epigenetic Regulation Studies: The utilization of DNA 6mA sequencing has proven instrumental in unlocking the intricate epigenetic landscape of organisms. This advanced method identifies 6mA modification sites throughout the genome, a crucial step in revealing their distribution patterns and their temporal dynamics. Our understanding of how 6mA modifications serve as regulatory elements in gene expression, chromatin conformation, and broader epigenetic mechanisms is crucial for deconstructing the sophisticated mechanisms governing diverse cellular activities and developmental trajectories.
Exploration of Disease Mechanisms: Disruptions in DNA 6mA modifications have been increasingly implicated in the pathology of an array of diseases, not least among them cancer, neurological dysfunctions, and metabolic disorders. The power of DNA 6mA sequencing lies in its ability to facilitate informed comparisons between pathological and healthy tissues, thereby pinpointing various 6mA modification patterns that reflect the afflicted disease states. Such telling findings expedite the identification of diagnostic biomarkers and possible therapeutic targets, and ultimately provide innovative directions for further disease prevention and control efforts.
Research in Developmental Biology: DNA N6-methyladenine (6mA) modifications play vital roles in a range of biological processes, including embryonic development, tissue differentiation, and organogenesis. The application of DNA 6mA sequencing allows scientists to catalog dynamic changes in 6mA distribution throughout various developmental stages, thereby enriching our understanding of the epigenetic control of developmental transitions. Unraveling the intersections between 6mA modifications and gene expression profiles can shed light on the molecular mechanisms steering organismal growth and intricate morphogenic events.
Studies on Environmental Responses: Environmental stimuli can influence the landscape of DNA 6mA modifications, contributing to an organism's adaptive response. Employing DNA 6mA sequencing enables the mapping of environmentally-induced 6mA profile alterations, thereby elucidating the adaptive roles of these epigenetic modifications in response to environmental perturbations. A thorough understanding of the molecular mechanisms underpinning these responses not only expands our comprehension of environmental adaptation but can also lend insights into the resilience of diverse biological systems.
Microbiome Investigation: DNA N6-methyladenine (6mA) modifications serve a pivotal role in the physiological dynamics, pathogenicity, and host-interactions of microbes. DNA 6mA sequencing permits the exhaustive profiling of these 6mA modifications within microbial genomes, therefore aiding the exploration of microbial epigenetic regulation and its consequential effects on microbial conduct and ecology. By discerning the function of 6mA modifications in microbial adaptability and pathogenesis, researchers have the potential to formulate innovative approaches for infectious disease control and the manipulation of microbial populations.
DNA 6mA Sequencing Workflow
DNA 6mA sequencing, a crucial tool in epigenetic research, involves several key steps to accurately identify and analyze N6-methyladenine (6mA) modifications in genomic DNA. Below is a typical DNA 6mA sequencing workflow:

Service Specifications
Our Methods of DNA 6mA Sequencing
Long read sequencing has contributed significantly to identify the genome-wide distribution of 6mA and can achieve single-nucleotide-resolution. As the absolute abundance of 6mA is relatively low, sufficient coverage higher than 100x is necessary. SMRT sequencing allows direct detection of modified nucleotides in the DNA template, including 6mA, 5mC, and 5-hydroxymethylcytosine, and does not adversely affect the determination of primary DNA sequence. Base modifications (6mA) are identified using RS Modification and Motif Analysis.
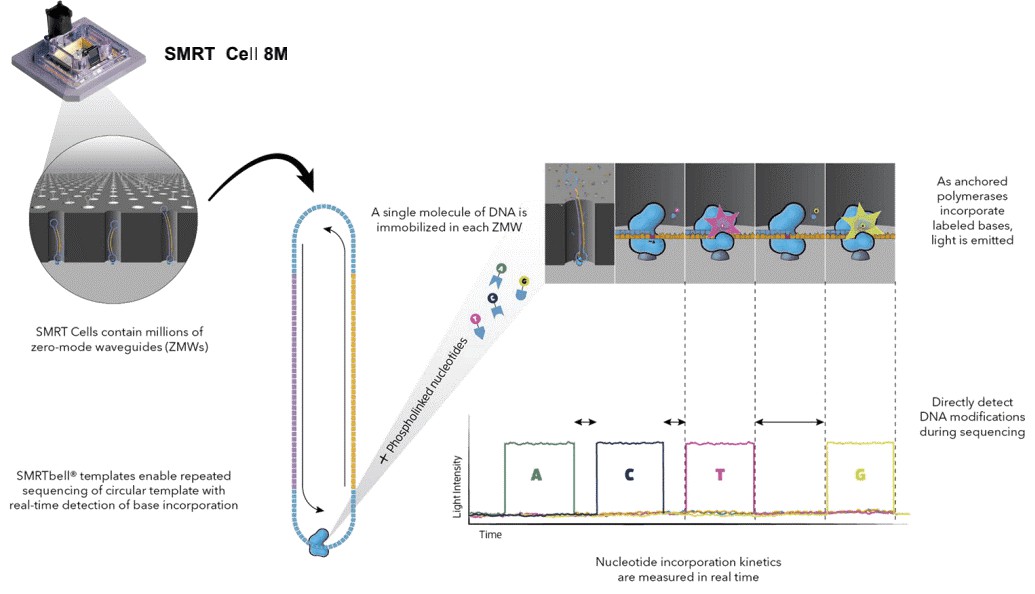 Figure 2. Simultaneous collection of data including DNA modifications.
Figure 2. Simultaneous collection of data including DNA modifications.
6mA DNA immunoprecipitation followed by deep sequencing (6mA DIP-Seq) was developed from existing approaches used to detect adenosine methylation in RNA. We developed this protocol and adapted it for the detection of 6mA in DNA. In this protocol, genomic DNA is extracted, fragmented and then DNA containing 6mA is pulled down with an antibody that recognizes 6mA in genomic DNA. After subsequent washes, DNA fragments that do not contain 6mA are eliminated, and the 6mA containing fragments are eluted from the antibody in order to be processed further for subsequent analyses.
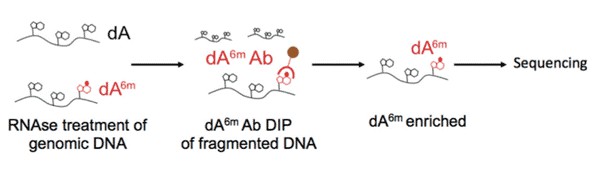 Figure 3. Illustration of 6mA DNA immunoprecipitation.
Figure 3. Illustration of 6mA DNA immunoprecipitation.
Sample Requirements
|
|
Sequencing
|
|
Data Analysis
We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in DNA 6mA sequencing for your writing (customization)
If you have additional requirements or questions, please feel free to contact us.
References:
- Koziol MJ, et al. Identification of Methylated Deoxyadenosines in Genomic DNA by dA6m DNA Immunoprecipitation. Bio Protoc. 2016 Nov 5;6(21).
- Chuan-Le Xiao, et al. Single-nucleotide-resolution sequencing of human N6-methyldeoxyadenosine reveals strand-asymmetric clusters associated with SSBP1 on the mitochondrial genome. Nucleic Acids Res. 2018 Dec 14;46(22):11659-11670.
- Li H, Zhang N, Wang Y, et al. DNA N6-methyladenine modification in eukaryotic genome. Frontiers in Genetics, 2022, 13: 914404.
- Li X, Zhang Z, Luo X, et al. The exploration of N6-deoxyadenosine methylation in mammalian genomes. Protein & cell, 2021, 12(10): 756-768.
Partial delivery chart examples:
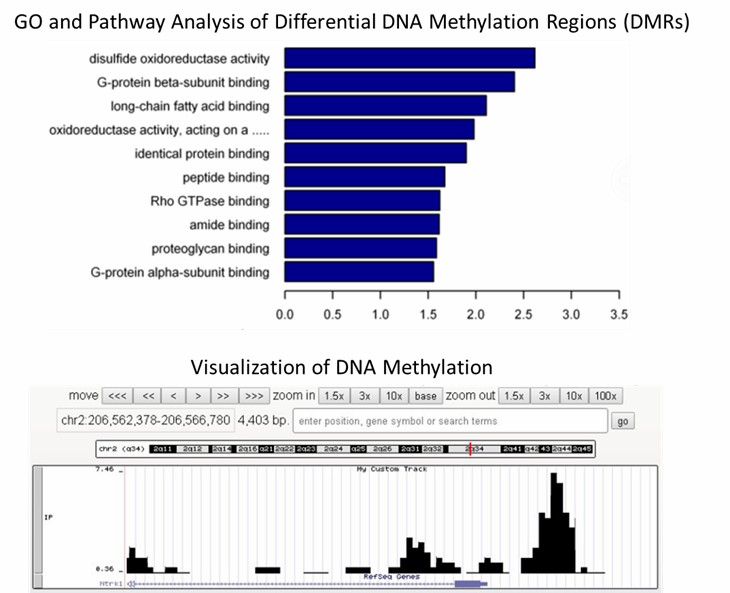
1. What are the most commonly used methods for DNA 6mA detection?
The methodologies commonly employed for the detection of DNA 6mA encompass a spectrum of techniques including liquid chromatography-tandem mass spectrometry (LC-MS/MS), antibody-dependent assays like immunoblotting and immunofluorescence (IF), immunoprecipitation coupled with sequencing (DIP-seq), and non-antibody-dependent approaches.
LC-MS/MS stands out for its remarkable sensitivity and specificity, enabling the precise quantification of DNA 6mA with sensitivity reaching up to one in ten million. This method aptly discriminates between DNA 6mA and RNA m6A or other DNA modifications. However, meticulous sample preparation is imperative to eradicate potential contamination, particularly from bacteria, which could yield false-positive outcomes.
Antibody-dependent techniques, such as immunoblotting and immunofluorescence, though less sensitive compared to LC-MS/MS, offer simplicity and widespread utility in probing 6mA. Nevertheless, the current antibodies lack discrimination between DNA 6mA and RNA m6A or m6Am, necessitating rigorous removal of RNA contamination to prevent spurious results.
DIP-seq emerges as a robust approach, amalgamating immunoprecipitation with sequencing, facilitating the precise detection and quantification of 6mA distribution across the genome. Nonetheless, optimization of antibody specificity for individual experimental systems is paramount, while the abundance of 6mA plays a pivotal role in successful enrichment.
Non-antibody-dependent methods play a pivotal role in validating antibody-dependent findings and mitigating nonspecific signals. One such method involves the utilization of methylation-sensitive restriction endonucleases to cleave DNA, yet its reliance on specific DNA motifs constrains its versatility. Another promising avenue is third-generation sequencing, exemplified by SMRT-seq, leveraging the kinetic properties of DNA polymerase during replication to detect 6mA. While SMRT-seq has proven successful in detecting 6mA in mammals and plants, challenges arise from the low abundance of 6mA, necessitating enrichment and robust sequencing coverage.
2. Which organisms contain 6mA?
The epigenetic mark, N6-methyladenine (6mA), is ubiquitously distributed across various life forms, most commonly occurring in prokaryotes, though also detected within certain eukaryotic species. Herein, we delineate certain organisms acknowledged for their possession of 6mA modifications:
In the realm of prokaryotes, 6mA modifications represent a predominant form of DNA modification, appearing extensively in varied bacterial species such as Escherichia coli and Clostridium perfringens, in addition to subsets of the archaea phylum, inclusive of Methanobacterium and Sulfolobus solfataricus.
Turning attention to eukaryotes, the discovery of 6mA modifications in this domain was somewhat deferred, yet recent studies have ascertained its existence in select organisms:
- Yeast: Certain yeast species, such as Saccharomyces cerevisiae, are known for featuring 6mA modifications.
- Nematodes: The eukaryotic organism, Caenorhabditis elegans, exhibits 6mA modifications.
- Fruit Flies: Evidence suggests a presence of 6mA modifications within the genome of Drosophila melanogaster.
- Plants: Various plants, such as Arabidopsis thaliana, exhibit 6mA modifications.
In line with advancements in sequencing technologies, the surveyance of 6mA modifications across larger spectra of eukaryotic organisms continues to intensify. It is expected that future discoveries will further augment the list of organisms known to harbor 6mA modifications.
3. How does DNA 6mA sequencing work?
DNA 6mA sequencing typically involves several steps: DNA extraction, fragmentation, enrichment of 6mA-modified DNA fragments, library preparation, high-throughput sequencing, and computational analysis. Various enrichment methods, such as antibody-based immunoprecipitation or chemical labeling, can be employed to selectively capture 6mA-modified DNA fragments prior to sequencing.
4. How is DNA 6mA sequencing data deciphered?
The analysis of DNA 6mA sequencing data generally encompasses preliminary processing of raw sequencing reads, alignment to the reference genome, identification of 6mA modification locales, differential examination between experimental conditions, and functional annotation of 6mA-customized regions. Numerous bioinformatic tools and software suites are available to facilitate each step in the analytical process.
5. What are the challenges associated with DNA 6mA sequencing?
Challenges in DNA 6mA sequencing include the efficient enrichment of 6mA-modified DNA fragments, the accurate detection of 6mA sites amidst other DNA modifications, and the interpretation of sequencing data to decipher the functional significance of 6mA modifications. Additionally, technical variability and bioinformatic complexities can pose challenges in data analysis and interpretation.
N6-methyladenine DNA modification in the human genome
Journal: Molecular cell
Impact factor: 15.280
Published: 19 July 2018
Backgrounds
Emerging from an extensive history of research on 5mC, largely buoyed by its widespread occurrence in eukaryotes, scholarly attention to 6mA has primarily been ensconced within prokaryotic boundaries. Be that as it may, recent strides in deep sequencing technologies have thrown open the doors to locate potential traces of 6mA in select eukaryotic domains, thereby sparking further inquiries into its consequential role in gene regulation and epigenetic inheritance. The examination referenced herein employed the precision of PacBio SMRT sequencing technology to lay bare the vestiges of 6mA in the human genome, with a particular emphasis on the DNA of mitochondria.
Methods
- DNA and Tissue samples
- Genome DNA extraction
- PacBio sequencing
- 6mA-IP-seq
- Measurement of the 6mA/A ratio by LC-MS/MS
- 6mA-IP-qPCR
- 6mA-RE-qPCR
- 6mA profiling
- Aligned
- Statistical enrichment of genes
Results
1. DNA 6mA Modification Occurs in the Human Genome
The study utilized PacBio sequencing data to identify over 880,000 6mA modification sites in the human genome, with a density of approximately 0.051% of total adenines. This density was found to be lower in humans compared to certain other organisms like fungi, Chlamydomonas, and C. elegans, but similar to that in Drosophila and pigs. Further validation using 6mA immunoprecipitation sequencing (6mA-IP-seq) and liquid chromatography-tandem mass spectrometry (LC-MS/MS) confirmed the presence of 6mA in human genomic DNA.
The distribution of 6mA sites across the human genome varied, with higher density observed in mitochondrial DNA compared to autosomal chromosomes. Chromosomes X and Y exhibited lower 6mA density, contrasting with the higher levels of DNA 5mC modifications. The study also identified differences in 6mA modification levels across different chromosomal regions, with enrichment observed in the middle modification level category. Additionally, the chromosomal distributions of 6mA-containing DNA fragments identified by different techniques were found to be consistent.
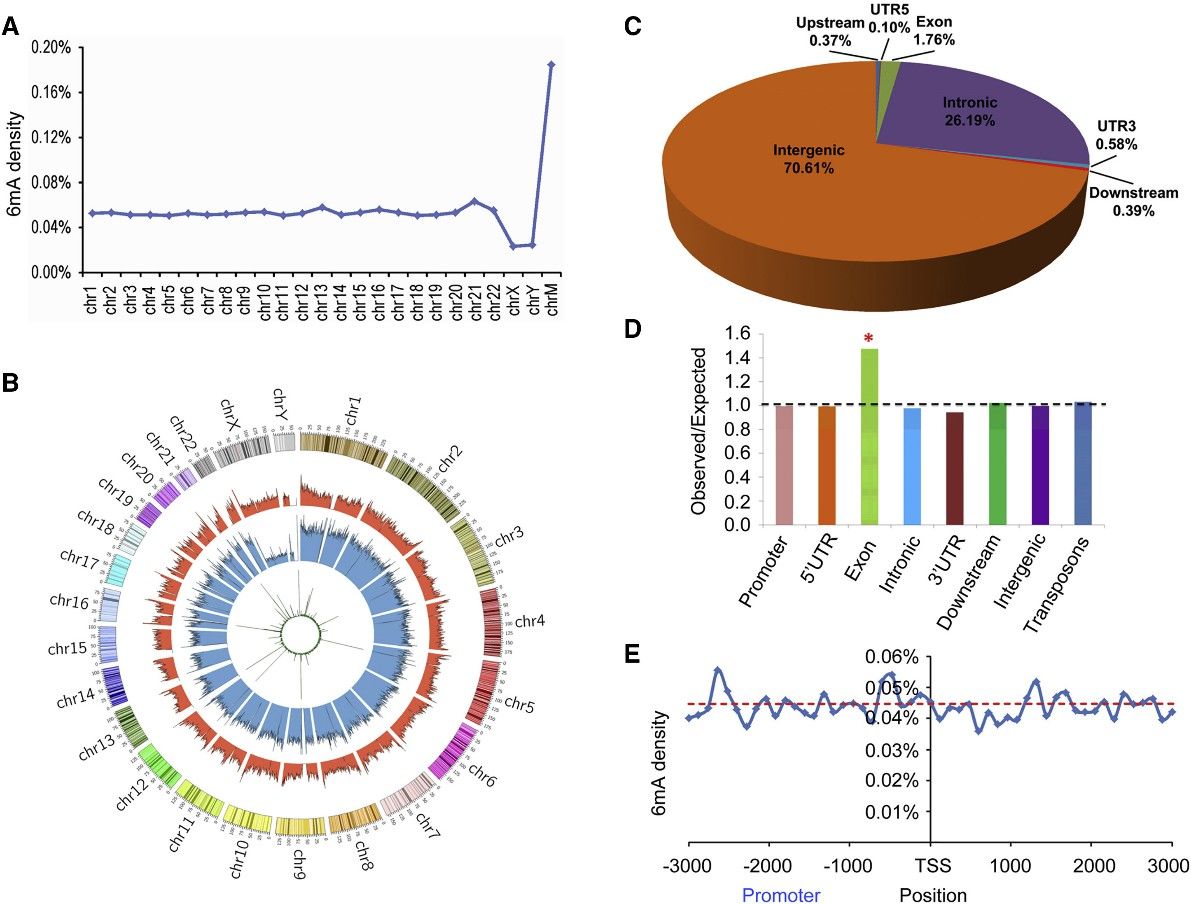 Figure 1. Distribution of 6mA Sites in the Human Genome.
Figure 1. Distribution of 6mA Sites in the Human Genome.
2. 6mA Is Significantly Enriched in Exon Regions
This intriguing research scrutinized the distribution of 6mA modifications across functional zones within the human genome, delineating that the preponderance of these sites were situated within intronic and intergenic regions. A substantial enrichment of 6mA was discerned within exon-coding domains, suggesting a putative role in gene expression modulation. This enrichment remained robust and consistent when analyzed via diverse sequencing techniques, demonstrating its independence from the variability in method sensitivity levels. However, in contrast to findings in other model organisms such as C. reinhardtii, there was an absence of discernible enrichment of 6mA around transcription initiation and termination sites, thus prompting further investigation in this field.
3. N6AMT1 Is a Methyltransferase for 6mA in the Human Genome
The study aimed to identify the methyltransferase responsible for DNA 6mA modification in humans. N6 adenine-specific DNA methyltransferase 1 (N6AMT1) was identified based on its similarity to bacterial 6mA DNA methyltransferases and its potential role as an S-adenosyl-l-methionine (AdoMet)-dependent methyltransferase. Silencing of N6AMT1 led to a decrease in DNA 6mA modification levels, while overexpression increased these levels in both in vivo and in vitro experiments. Recombinant N6AMT1-Flag efficiently increased 6mA modification levels in synthetic oligonucleotide substrates in vitro, an effect abolished by mutation of the catalytic conserved motif NPPY. Furthermore, knockout of N6AMT1 resulted in reduced genomic DNA 6mA levels, which were rescued by ectopic expression of wild-type N6AMT1 but not a mutant lacking the NPPY motif. These findings collectively establish N6AMT1 as the methyltransferase responsible for DNA 6mA modification in the human genome.
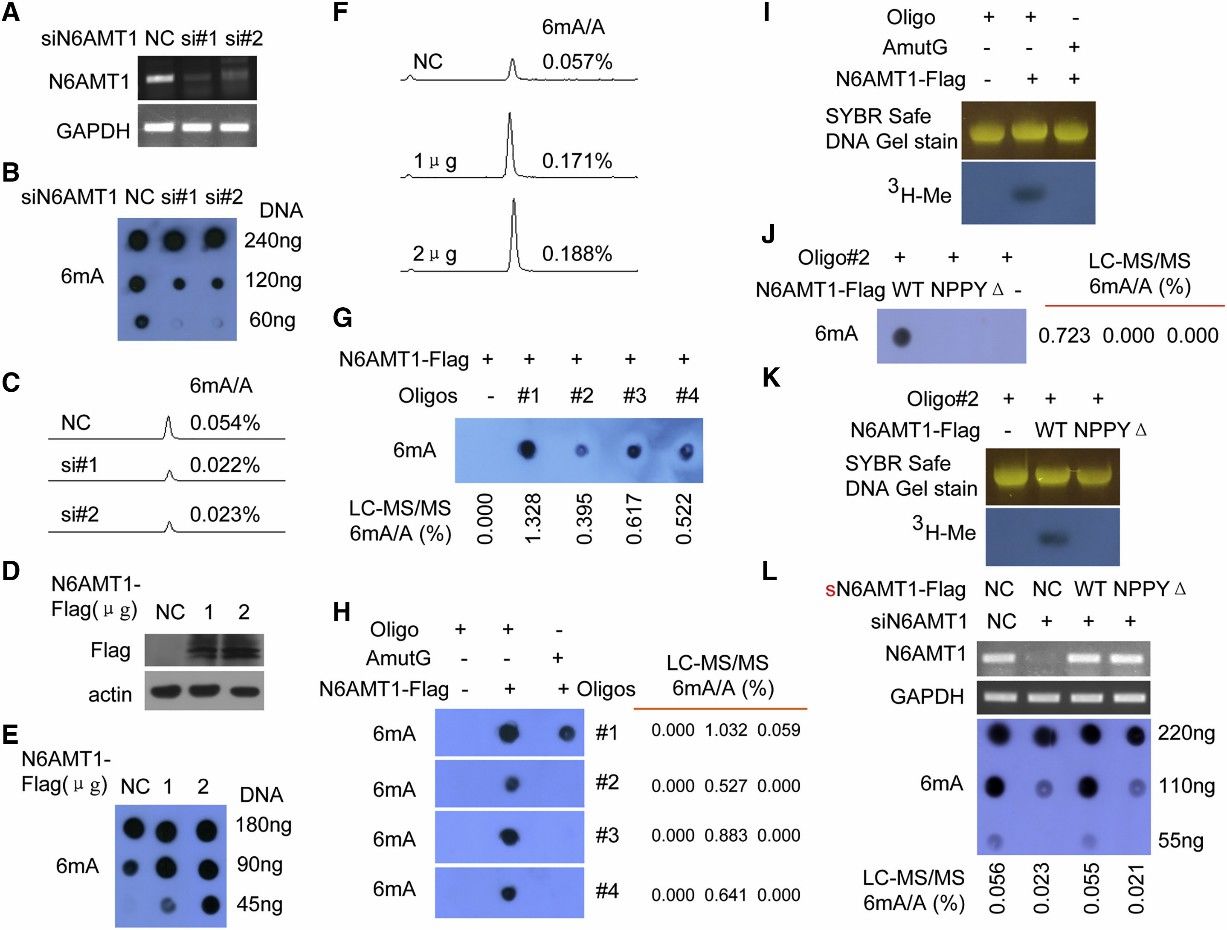 Figure 2. N6AMT1 Is a Methyltransferase for DNA 6mA Methylation in Human, and NPPY Is the Catalytic Motif of N6AMT1
Figure 2. N6AMT1 Is a Methyltransferase for DNA 6mA Methylation in Human, and NPPY Is the Catalytic Motif of N6AMT1
4. Decrease of Genomic DNA 6mA Levels Promotes Tumorigenesis
The scientific inquiry delved into the implications of DNA 6mA modification on tumorigenesis by modulating the expression of N6AMT1 and ALKBH1 within malignant cells. An intriguing consequence of N6AMT1 suppression, which resulted in diminished cellular genomic DNA 6mA concentrations, was the promotion of tumorigenic properties such as cell proliferation, colony origination, migration, and intrusion; whereas an upregulated N6AMT1 counteracted these processes. Contrarily, the suppression of ALKBH1, causing an augmentation in cellular genomic DNA 6mA concentrations, impeded these tumorigenic properties, which were again felt in a reversed manner upon ALKBH1 overexpression. In vivo validations converged with these observations, evidenced by accelerated tumour progression in xenografts hosting N6AMT1-suppressed cells, and a converse tumor progression trend in those hosting ALKBH1-suppressed cells. Metastatic nodules manifested within murine lungs mirrored the N6AMT1 and ALKBH1 expression landscapes. The forecast of cancer-related mortality risk, as measured by Kaplan-Meier survival analysis, was augmented in mice bearing N6AMT1-suppressed xenografts, while it showed a decrease in those bearing ALKBH1-suppressed xenografts. Findings from rescue assays employing wild-type and mutant avatars of N6AMT1 and ALKBH1 reinforced these observations. Collectively, these findings point to an enlightening hypothesis wherein diminished genomic DNA 6mA modification within human cancer cells has the potential to amplify tumorigenesis.
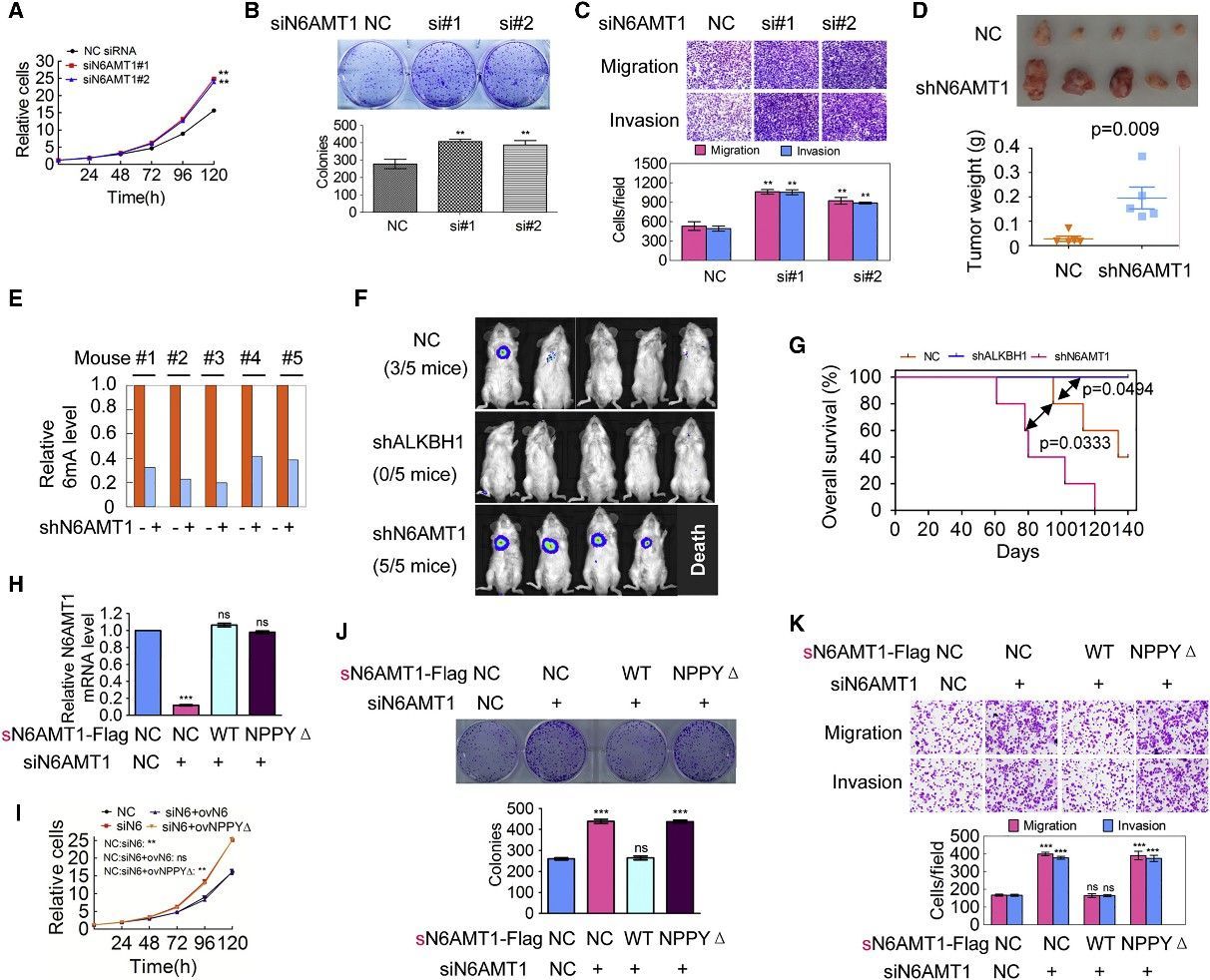 Figure 3. Decrease of Genomic 6mA Modification Level Promotes Tumorigenesis
Figure 3. Decrease of Genomic 6mA Modification Level Promotes Tumorigenesis
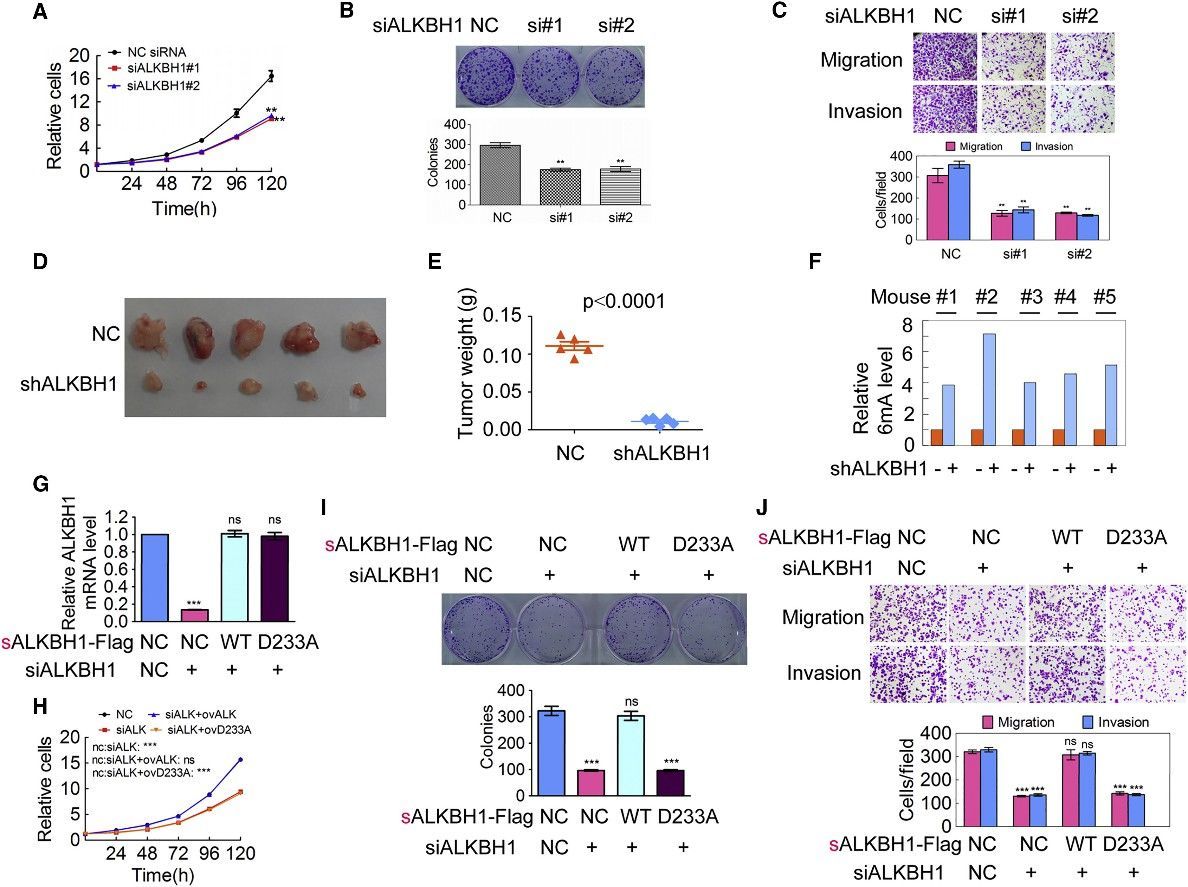 Figure 4. Increase of Genomic 6mA Modification Level Inhibits Tumorigenesis
Figure 4. Increase of Genomic 6mA Modification Level Inhibits Tumorigenesis
Conclusion
The scholarly pursuit in question strove to discern the occurrence and relevance of DNA N6-methyladenine (6mA) amendments in human cells, primarily considered as a phenomenon characteristic of prokaryotes prior to this study. Through the detection of over 880,000 6mA sites encompassing the human genome and conducting a comprehensive evaluation of their distribution and functional implications, the research was successful in unearthing the role 6mA plays in gene regulation. Notably, the enzymes coined as N6AMT1 and ALKBH1 surfaced as integral contributors in administrating the dynamism of 6mA modifications. Of critical importance was the finding that fluctuations in 6mA levels were inherently tied to the genesis of cancer, which suggested the potential value of 6mA as a biomarker and potential therapeutic fulcrum in the management of cancerous ailments.
Reference:
- Xiao C L, Zhu S, He M, et al. N6-methyladenine DNA modification in the human genome. Molecular cell, 2018, 71(2): 306-318. e7.


 Sample Submission Guidelines
Sample Submission Guidelines