MicroRNA (miRNA) expression profiling via microarrays constitutes a sophisticated method for elucidating gene regulation at the post-transcriptional level. This technique enables researchers to examine miRNA expression patterns across diverse biological conditions and species, thereby offering profound insights into various cellular processes and disease mechanisms. CD Genomics offers a comprehensive miRNA microarray profiling service, utilizing cutting-edge platforms renowned for their exceptional accuracy and sensitivity.
What is Expression Profiling with Microarrays
Expression profiling with microarrays is a sophisticated high-throughput technique that enables the simultaneous measurement of expression levels for thousands of genes. This method involves hybridizing RNA samples to a microarray chip embedded with complementary probes. By capturing and quantifying the hybridization signals, researchers can assess gene expression across a wide range of conditions. This approach is instrumental for identifying genes with differential expression, elucidating complex regulatory networks, and investigating the molecular mechanisms underlying various biological processes.
How to Do miRNA Profiling
MicroRNA profiling is essential for deciphering gene regulation and elucidating disease mechanisms. Below is a streamlined overview of the methodologies employed:
1. Traditional Methods
- Northern Blotting: This classical approach detects miRNAs through probe hybridization. While reliable, it is characterized by low sensitivity, requires substantial RNA quantities, and is time-intensive.
- Microarray: This technique enables high-throughput detection by hybridizing miRNAs with DNA probes on a chip. Advances such as LNA-modified probes and nanoparticle-based enhancements have significantly improved its sensitivity and specificity.
- qRT-PCR: Considered the gold standard, qRT-PCR offers exceptional sensitivity and specificity. Techniques such as tailing and stem-loop RT-PCR address the challenges posed by miRNA's short sequences, with TaqMan probes providing superior specificity, albeit at a higher cost compared to SYBR Green methods.
- Droplet Digital PCR (ddPCR): A newer advancement, ddPCR allows for absolute quantification of miRNAs with high sensitivity and precision. It involves partitioning samples into individual droplets and quantifying miRNAs without reliance on standard curves.
2. Emerging Methods
- Nanomaterial-Based Detection: This approach leverages nanomaterials such as gold nanoparticles and quantum dots to enhance detection capabilities. Techniques like colorimetric assays and fluorescence-based methods offer heightened sensitivity and reduced sample requirements.
What is miRNA Microarray
miRNA microarrays are advanced analytical tools designed specifically to detect and quantify miRNA expression profiles. Unlike conventional gene expression microarrays, which target mRNA, miRNA microarrays employ probes that are specific to miRNA sequences. This enables the simultaneous detection of thousands of miRNAs, offering a comprehensive overview of miRNA expression across various conditions.
The prominence of miRNA expression profiling stems from the understanding that miRNAs highly expressed in particular tissues, cell types, or developmental stages often play crucial regulatory roles. For accurate and efficient analysis, it is advisable to engage with specialists in miRNA and microarray technologies. These experts operate in state-of-the-art laboratories that adhere to stringent quality control protocols and deliver rapid results. They offer sophisticated data analysis tailored to meet the specific needs of your research, ensuring both high-quality outcomes and adherence to budgetary constraints.
Advantages and Features of miRNA Microarray
- High Throughput: miRNA microarrays can detect thousands of miRNAs simultaneously, providing an efficient way to profile miRNA expression comprehensively.
- Comprehensive Coverage: They cover a wide range of known miRNAs, detecting both abundant and low-abundance miRNAs for thorough expression analysis.
- Reproducibility: Microarrays offer consistent and reliable results, allowing accurate comparisons of miRNA expression across different experiments and conditions.
- Cost-Effectiveness: Compared to RNA sequencing, miRNA microarrays are more economical for large-scale profiling, offering good value for extensive studies.
- High Sensitivity: These microarrays are designed to detect miRNAs at low concentrations, ensuring precise and accurate measurement of expression levels.
- Complete content flexibility (all species)
- Optimized RNA hybridization probes
- Cost-effective, one-stop solution
- Experienced service provider handling various research & clinical sample types (including FFPE and Blood/Plasma Samples)
Applications of miRNA Microarray
- To distinguish expression signatures associated with diagnosis, prognosis and therapeutic interventions.
- Conducting a genome-wide analysis of miRNA expression of normal and disease samples, such as cancer.
- Profiling miRNA expression reveals changes associated with diseases like cardiovascular conditions, neurological disorders, and metabolic syndromes, which can help identify potential therapeutic targets.
- In drug discovery, miRNA microarrays assist in pinpointing miRNA targets and assessing the effects of drug candidates on miRNA expression, facilitating the creation of innovative therapies.
- Genome-wide analysis of miRNA expression in normal and diseased samples, such as cancer, aids in distinguishing expression profiles relevant to diagnosis, prognosis, and treatment responses.
miRNA Microarray Workflow
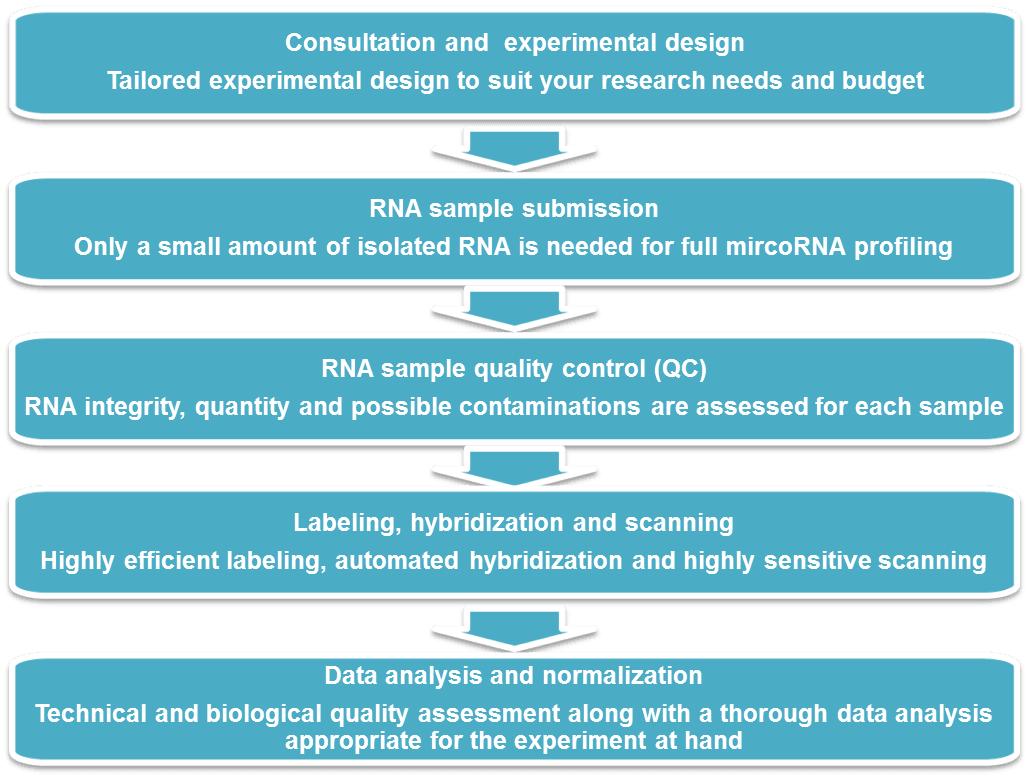
Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategy
|
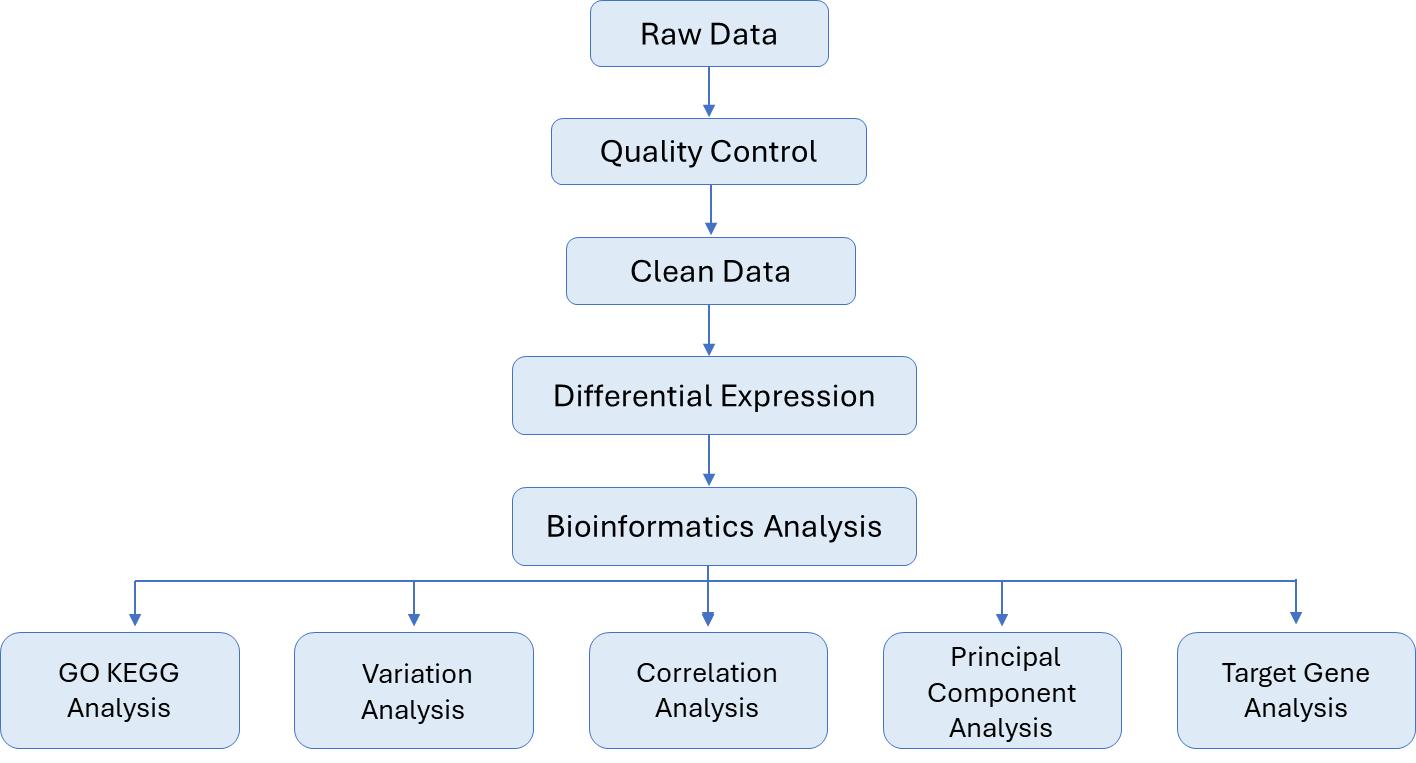
| Bioinformatics Analysis We provide multiple customized bioinformatics analyses:
|
Recommendations and Custom Service
Table 1 Agilent miRNA Expression Arrays
| Microarray | Format(s) | Detected miRNAs | Database |
| Human miRNA Microarray V19.0 | 8 х 60K | 2006 | Sanger miRBase V19.0 |
| Mouse miRNA Microarray V19.0 | 8 х 60K | 1247 | Sanger miRBase V19.0 |
| Rat miRNA Microarray V19.0 | 8 х 15K | 719 | Sanger miRBase V19.0 |
Table 2 Affymetrix miRNA Expression Arrays
| Microarray | Detected miRNAs | Database |
| GeneChip® miRNA 4.0 Array | mature miRNAs and Pre-miRNA can be detected for humans, mouse, and rat. Human snoRNA and scaRNA can also be detected | Sanger miRBase V17.0 |
Table 3 TaqMan® Array MicroRNA Cards
| Microarray | Product Size | Detected miRNAs | Database |
| TaqMan® Array Human MicroRNA A+B Cards Set v3.0 |
8 pack | 757 One negative Control |
Sanger miRBase V20.0 |
| TaqMan® Array Rodent MicroRNA A+B Cards Set v3.0 |
8 pack | 644 (mouse) 376 (rat) One negative Control |
Sanger miRBase V20.0 |
Analysis Pipeline
Deliverables
- The original and processed microarray scan images
- An array layout file
- A raw intensity data file in Excel
- A fully processed data file in Excel
- A list of up and down regulated transcripts that are called based on a statistical analysis
- Additionally, for each batch of samples, the customer receives a data summary containing a catalog of data files, images of representative regions of corresponding arrays, and descriptions of specific features of the arrays.
CD Genomics can also help create your custom miRNA microarray. We are ready to help you with your custom array needs, whether it's a standard design or something more creative.
For details, please feel free to contact us with any questions by completing a no-obligation Quote Request.
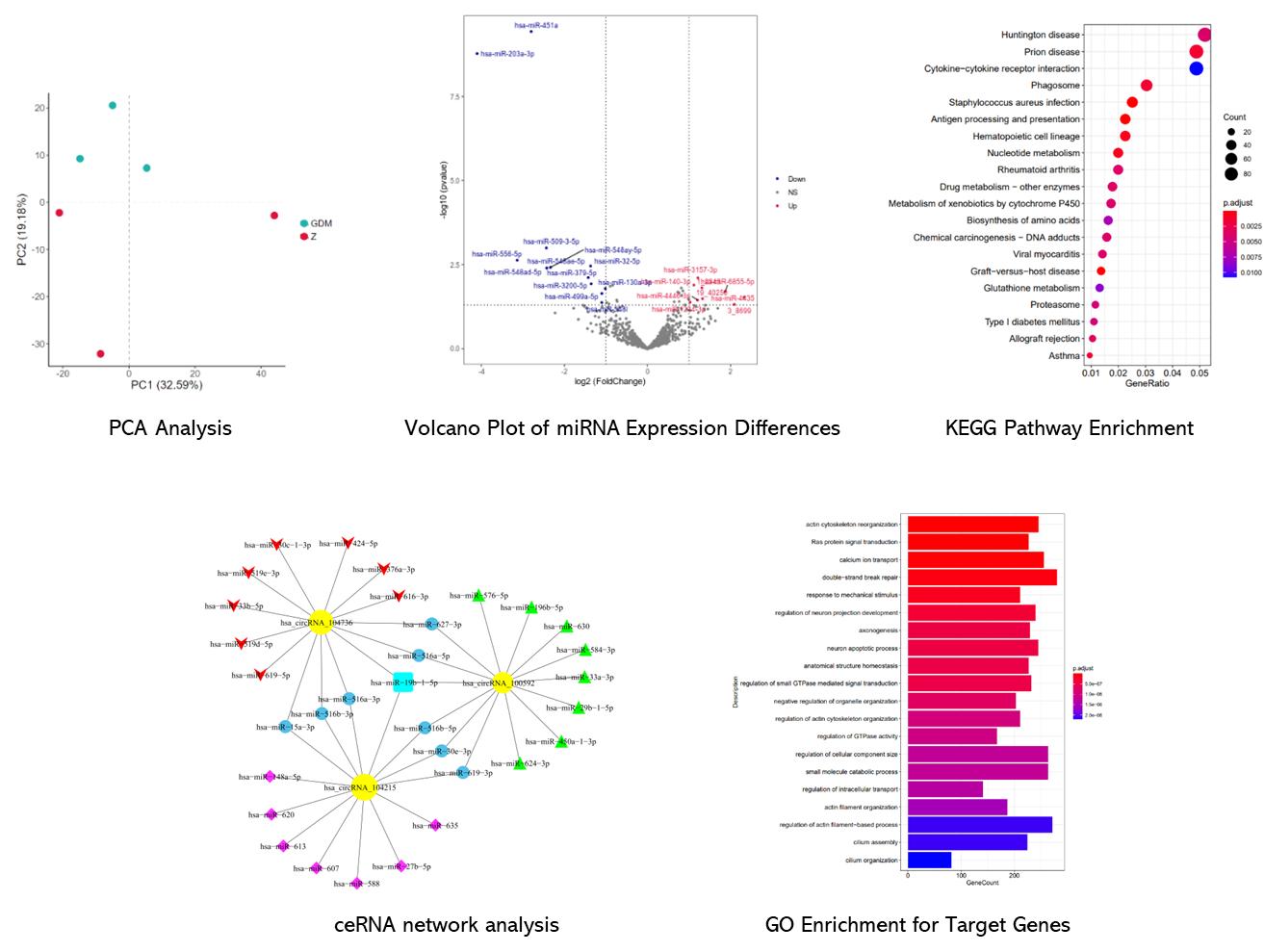
Partial results are shown below:
1. What types of samples can be used for miRNA microarray profiling?
miRNA microarray profiling can be performed using various sample types, including tissues, cells, and biofluids such as plasma or serum. It is crucial to use RNA extraction methods that preserve small RNAs to ensure accurate miRNA profiling.
2. How long does a miRNA microarray experiment take?
The duration of a miRNA microarray experiment, encompassing sample preparation through to data analysis, generally ranges from several days to a few weeks. This timeframe is influenced by the complexity of the experimental setup and the number of samples processed.
3. Are miRNA microarrays suitable for all types of miRNA studies?
miRNA microarrays are well-suited for investigations involving known miRNAs and are effective for applications such as biomarker discovery and functional studies. However, they may not detect all novel or less-characterized miRNAs.
4. How do miRNA microarrays compare to RNA sequencing?
While RNA sequencing provides a more exhaustive analysis and identifies novel miRNAs, miRNA microarrays are a cost-effective, high-throughput option that delivers robust data for known miRNAs.
LncRNA-miRNA-mRNA expression variation profile in the urine of calcium oxalate stone patients
Journal: BMC Medical Genomics
Impact Factor: 3.622
Published: 29 April 2019
Background
Urolithiasis, especially calcium oxalate (CaOx) stones, is common and problematic, with high recurrence rates. Factors like hyperoxaluria and inflammation contribute to stone formation. MicroRNAs and long-non-coding RNAs play roles in regulating gene expression and might influence stone formation. This study analyzes urine samples from urolithiasis patients to explore the interactions between miRNAs, mRNAs, and lncRNAs, using microarray technology for a comprehensive understanding.
Materials & Methods
Sample Preparation
- Calcium oxalate stone patients
- Urine
- RNA extraction
Method
- Microarray analysis
- Agilent miRNA Microarrays 8x60K
- Construction of lncRNA-miRNA-mRNA
- Functional analysis
- Statistical analysis
Results
In urine samples from CaOx kidney stone patients, the authors identified nine miRNAs with significant changes, including four that were upregulated and five downregulated. Additionally, analysis revealed alterations in 883 mRNAs and 1002 lncRNAs, with many either upregulated or downregulated in the stone group compared to controls. The authors constructed a ceRNA network to explore interactions among these differentially expressed RNAs, highlighting key regulatory relationships and pathways involved in CaOx stone formation, including ROS production and various metabolic processes.
Table 1. Total differentially expressed miRNAs between stone-forming and normal group
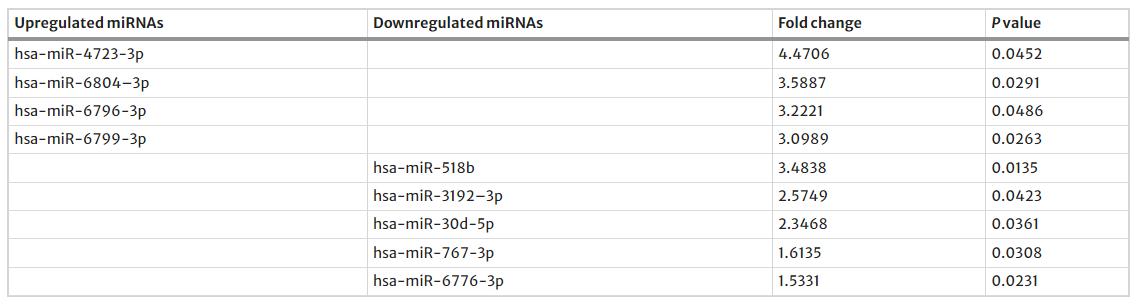
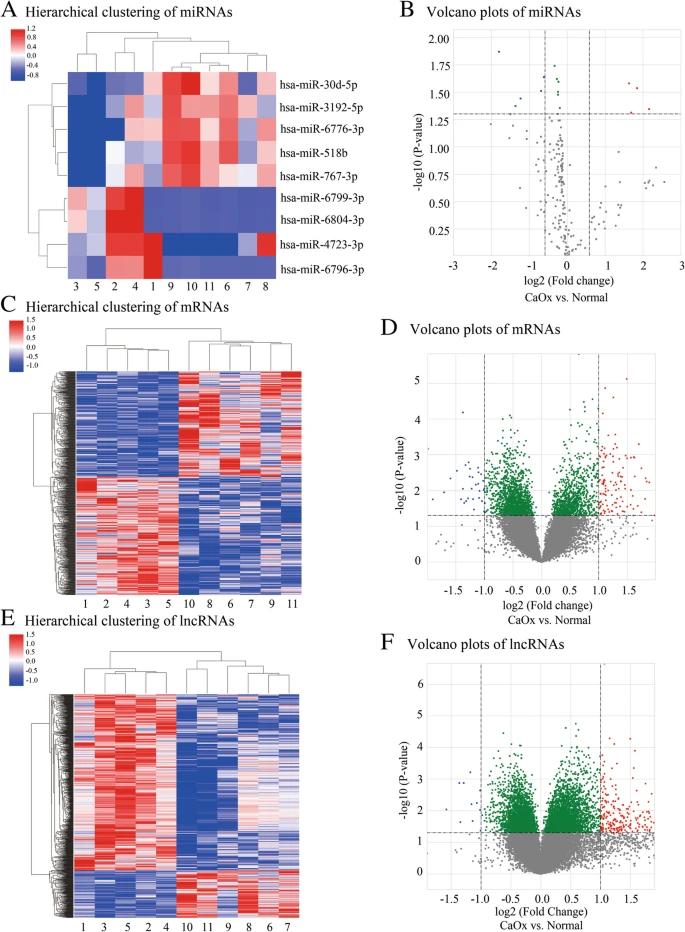 Fig 1. Hierarchical clustering and volcano plots of dysregulated miRNA, mRNA and lncRNA.
Fig 1. Hierarchical clustering and volcano plots of dysregulated miRNA, mRNA and lncRNA.
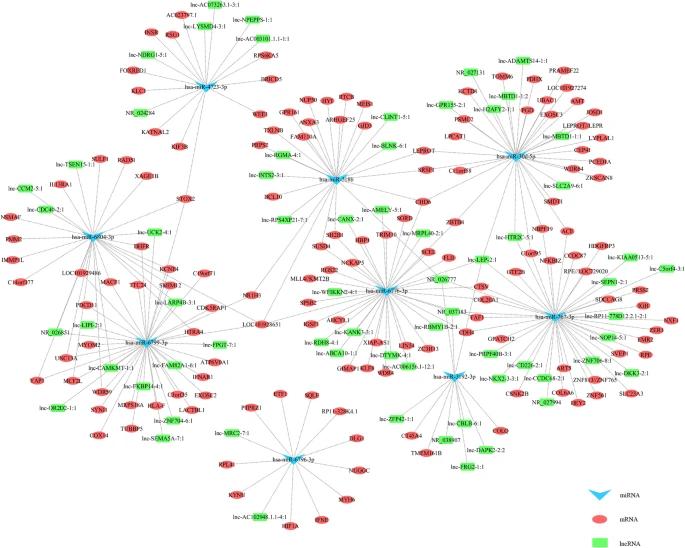 Fig 2. CeRNA network responded to the miRNA, mRNA and lncRNA variations in urine of CaOx stone patients.
Fig 2. CeRNA network responded to the miRNA, mRNA and lncRNA variations in urine of CaOx stone patients.
Conclusion
The study clearly defined miRNA, mRNA, and lncRNA expression differences in urine between CaOx stone patients and healthy individuals, using GO and KEGG analyses to uncover relevant gene functions and pathways. This work could lead to new diagnostic biomarkers or treatment targets for CaOx kidney stones and guide future research on urolithiasis mechanisms.
Reference
- Liang X, Lai Y, Wu W, et al. LncRNA-miRNA-mRNA expression variation profile in the urine of calcium oxalate stone patients. BMC Medical Genomics. 2019, 12:1-1.
Here are some publications that have been successfully published using our services or other related services:
Use of biostimulants for water stress mitigation in two durum wheat (Triticum durum Desf.) genotypes with different drought tolerance
Journal: Plant Stress
Year: 2024
The Restriction-Modification Systems of Clostridium carboxidivorans P7
Journal: Microorganisms
Year: 2023
In the land of the blind: Exceptional subterranean speciation of cryptic troglobitic spiders of the genus Tegenaria (Araneae: Agelenidae) in Israel
Journal: Molecular Phylogenetics and Evolution
Year: 2023
Genetic Modifiers of Oral Nicotine Consumption in Chrna5 Null Mutant Mice
Journal: Front. Psychiatry
Year: 2021
A high-density genetic linkage map and QTL identification for growth traits in dusky kob (Argyrosomus japonicus)
Journal: Aquaculture
Year: 2024
Genomic and chemical evidence for local adaptation in resistance to different herbivores in Datura stramonium
Journal: Evolution
Year: 2020
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
