CD Genomics provides a comprehensive ChIP-on-chip service, encompassing all stages from initial sample preparation to final data delivery. Available to both academic and commercial researchers, this service leverages highly esteemed microarray platforms, ensuring the production of high-quality and reliable results.
What is the ChIP-chip Technique
Chromatin Immunoprecipitation (ChIP) has emerged as a pivotal method for in vivo analysis of protein-DNA interactions, evolving over the past decade into a cornerstone of epigenetic research. The technique, also known as site-specific analysis, saw significant advancements with its integration into chip technology, giving rise to Chromatin Immunoprecipitation-chip (ChIP-chip or ChIP-on-chip). It is crucial to distinguish the capitalization in ChIP-chip, where 'ChIP' denotes chromatin immunoprecipitation and 'chip' signifies gene chip technology.
The fundamental principle of ChIP-chip involves fixing protein-DNA complexes in live cells, fragmenting them into small chromatin fragments within a defined size range, and immunoprecipitating these complexes using specific antibodies. This process enriches DNA segments bound by the target protein, enabling purification and detection to reveal crucial protein-DNA interaction details.
A notable distinction from traditional ChIP lies in ChIP-on-chip's ability to identify potential target genes bound by a given protein A without prior knowledge of specific DNA sequences. This approach captures a comprehensive set of DNA fragments associated with protein A, facilitating the downstream screening and identification of its regulatory targets.
What is ChIP-on- chip Used For
ChIP-on-chip is a robust method used to pinpoint genomic locations where specific proteins interact with DNA, offering critical insights into gene regulation and chromatin dynamics.
- Identification of Protein-DNA Interactions: ChIP-on-chip precisely identifies genomic regions where proteins bind, essential for unraveling how transcription factors and chromatin modifiers orchestrate gene regulation.
- Mapping Regulatory Elements: It characterizes enhancers, silencers, and other non-coding regions, unveiling their pivotal roles in governing gene expression patterns and influencing cellular differentiation processes.
- Study of Epigenetic Modifications: This technique scrutinizes histone modifications such as acetylation and methylation, providing key insights into chromatin structure dynamics and their impact on gene regulation in various developmental stages and diseases.
- Insights into Disease Mechanisms: ChIP-on-chip correlates protein-DNA interactions with disease pathways, facilitating the discovery of biomarkers and fostering the development of targeted therapeutic interventions.
- Integration with Systems Biology: Data derived from ChIP-on-chip aids in constructing comprehensive regulatory networks, deepening our understanding of intricate biological systems and predicting cellular responses across different conditions.
- Advancing Precision Medicine: By pinpointing genetic variations that affect protein-DNA interactions, ChIP-on-chip enables personalized therapeutic strategies tailored to individual genomic profiles, thus optimizing treatment outcomes in precision medicine.
Difference Between ChIP-on-chip and ChIP-seq
ChIP-on-chip and ChIP-seq, though both designed to probe protein-DNA interactions, diverge remarkably in their methodologies and applications. ChIP-on-chip employs the hybridization of immunoprecipitated DNA fragments onto a microarray platform. This approach allows for the concurrent examination of multiple genomic loci, making it particularly suited for broad, expansive genomic screenings.
In stark contrast, ChIP-seq utilizes high-throughput sequencing technology to directly sequence the immunoprecipitated DNA fragments. This method offers unparalleled single-base resolution and heightened sensitivity, enabling precise identification of protein-DNA binding sites. The meticulous detail in mapping these interactions provided by ChIP-seq surpasses what ChIP-on-chip can deliver.
Consequently, while ChIP-on-chip shines in its ability to efficiently screen large genomic regions, ChIP-seq has become the gold standard for detailed and accurate delineation of protein-DNA interactions. Each technique, therefore, extends unique advantages tailored to the specific aims and scope of the research being conducted.
Advantages of ChIP-on-chip Service
- Comprehensive genome-wide and focused coverage
- Superior microarray performance and capability
- Greater confidence in binding event data
- Flexible and versatile user-defined microarray formats
- Greater confidence in binding event data
- Quick turnaround
- Easy-to-use data analysis software
- Access to probe sequence and annotation information
Applications of Our ChIP-on-chip Service
- Uncover and validate gene regulation and regulatory networks by comprehensive determination of promoter occupancy.
- Identify and characterize molecular events associated with processes for transcription, DNA replication and repair, as well as with chromatin modifications and DNA methylation.
- Elucidate modes of action and potential therapeutic activities of compounds and target genes by mapping gene regulatory networks relevant to disease and pathophysiological states.
- Validate and augment existing gene expression data with authentic binding events.
- Identify, assess, and monitor biomarkers response to protein-DNA binding events to serve as bioassays or toxicant signatures for toxicogenomics.
- Uncover and profile off-target events as well as validate primary and secondary effects in screening of candidate compounds, siRNAs, therapeutics, etc.
Our ChIP-on-chip Service Workflow
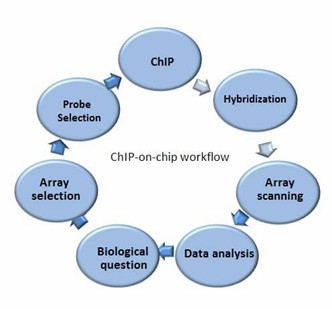
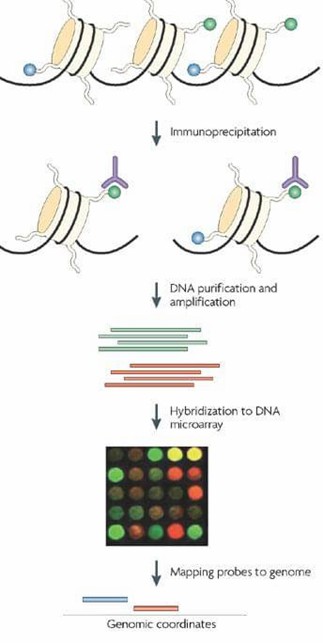
Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategy
|
| Bioinformatics Analysis We provide multiple customized bioinformatics analyses:
|
Recommendations and Custom Service
Table 1 Agilent ChIP-on-chip Arrays
| Microarray | Species | Format(s) | Coverage |
| SurePrint G3 Human Promoter Array | Human | 1 х 1M | 21,000 RefSeq transcripts |
| SurePrint G3 Mouse Promoter Array | Mouse | 1 х 1M | 19,000 RefSeq transcripts |
| Others (please click here ) | - | - | - |
Table 2 Affymetrix ChIP-on-chip Arrays
| Microarray | Species | Unite size | Coverage |
| Human Promoter 1.0R Array | Human | 2 arrays | 25,500 RefSeq promoters (-2.45 kb~ +2.75 kb) |
| Mouse Promoter 1.0R Array | Mouse | 2 arrays | 28,000 RefSeq promoters (-2.5 kb~ +6 kb) |
| Others | - | - | - |
Analysis Pipeline
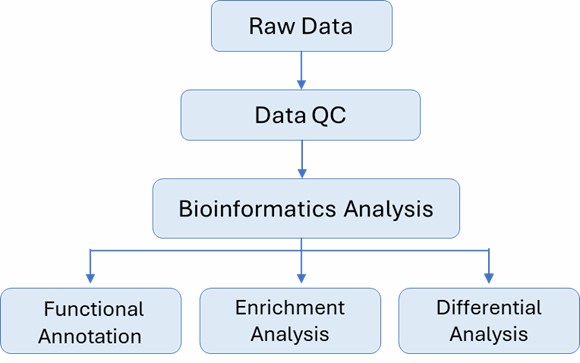
Deliverables
- Raw data
- Experimental results
- Data analysis report
CD Genomics can also help create your own custom microarray. We are ready to help you with your custom array needs, whether it's a standard design or something more creative.
For more details, please feel free to contact us with any questions at any time by completing a no obligation quote request.
Partial results are shown below:
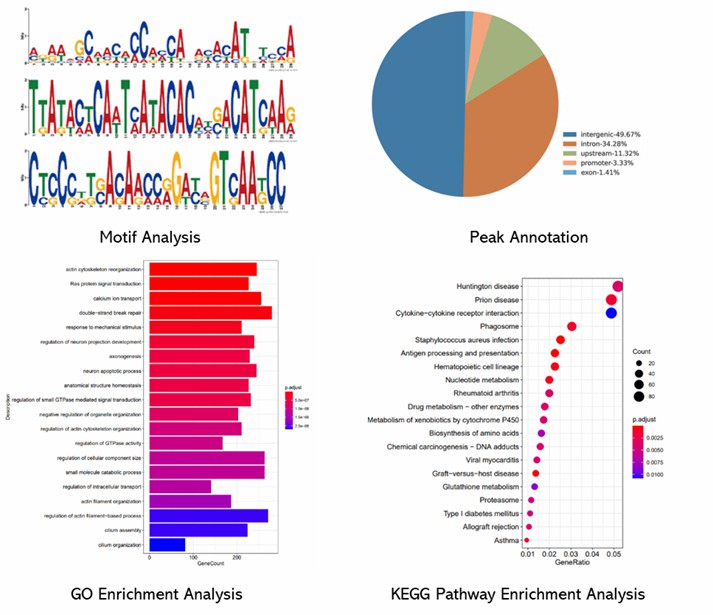
1. How sensitive is ChIP-on-chip in detecting protein-DNA interactions?
ChIP-on-chip demonstrates high sensitivity and is proficient at detecting protein-DNA interactions, even when these occur at low abundance. This capacity renders it a versatile technique for the investigation of both common and rare binding events.
2. How can ChIP-on-chip data be analyzed?
Analyzing ChIP-on-chip data involves several nuanced steps. Initially, raw microarray data must be meticulously processed to identify enriched DNA fragments. Following this, bioinformatics tools are employed to map these fragments to the genome, annotating them with nearby genes and regulatory elements. Statistical methods, particularly peak calling algorithms, are crucial in determining significant binding sites. Finally, the integration of ChIP-on-chip data with other omics datasets enables thorough analyses of regulatory networks and functional annotations.
3. What are the limitations of ChIP-on-chip?
While ChIP-on-chip has distinct advantages, it is not without limitations. The technique requires prior knowledge of potential binding sites and is reliant on the quality and availability of microarray platforms. Data accuracy can be affected by issues such as cross-hybridization and background noise from non-specific binding. Moreover, ChIP-on-chip may fail to capture transient or weak protein-DNA interactions effectively, thus limiting its applicability in the study of certain dynamic biological processes.
Interplay of miR-137 and EZH2 contributes to the genome-wide redistribution of H3K27me3 underlying the Pb-induced memory impairment
Journal: Cell Death & Disease
Impact factor: 5.959
Published: 11 September 2019
Background
This study links developmental lead (Pb) exposure to impaired spatial memory through epigenetic changes involving H3K27me3 and EZH2. Pb reduces H3K27me3 levels by suppressing EZH2 expression early on, affecting memory in rats. EZH2 overexpression restores H3K27me3 and improves memory. MiR-137 and EZH2 regulate H3K27me3 levels in response to Pb. ChIP-chip studies show Pb alters H3K27me3 distribution, impacting pathways like transcriptional regulation and Wnt signaling, crucial in neurodegenerative diseases influenced by environmental factors.
Materials & Methods
Sample Preparation
- Rat brains
- Neuronal cells
- RNA extraction
Method
- Quantitative RT-PCR analysis
- co-IP
- ChIP-chip
- miRNA profiling
- Statistical analysis
- ChIP-chip peaks annotation
Results
Pb induces genome-wide changes in H3K27me3 occupancy: ChIP-chip analysis revealed that Pb treatment alters H3K27me3 binding across the genome, with 327 genes gaining and 261 genes losing H3K27me3 marks. These changes affect pathways crucial for neuronal function and development, highlighting Pb's broad impact on epigenetic regulation in neurons.
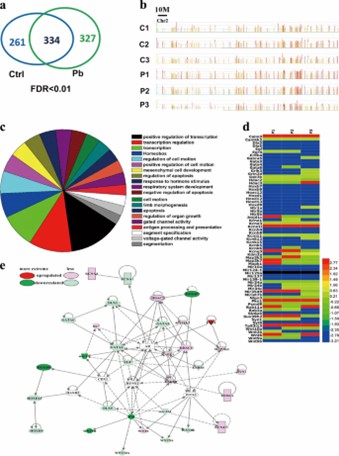 Fig. 1: Genome-wide redistribution of H3K27me3 is elicited in hippocampal neurons by Pb exposure.
Fig. 1: Genome-wide redistribution of H3K27me3 is elicited in hippocampal neurons by Pb exposure.
Pb modulates the bivalent state of the Wnt9b locus: Pb exposure reduces H3K27me3 enrichment at the Wnt9b promoter, leading to increased Wnt9b expression. This locus also shows bivalent regulation with H3K4me3, implicating Pb in the disruption of histone modifications associated with gene activation and repression, potentially contributing to neurodevelopmental abnormalities induced by Pb exposure.
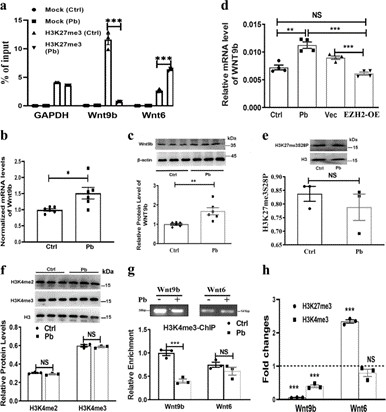 Fig. 2: H3K27me3 targets Wnt9b in a bivalent manner.
Fig. 2: H3K27me3 targets Wnt9b in a bivalent manner.
Conclusion
This study elucidates how lead (Pb) disrupts synaptic function, glutamate metabolism, and neuronal pathways, contributing to memory deficits. Targeting H3K27me3 presents a promising avenue for alleviating Pb-induced memory impairments, offering potential therapeutic implications for neurodegenerative diseases.
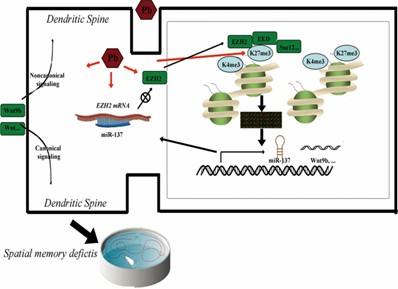 Fig. 3: Proposed model of H3K27me3's roles in spatial memory deficits caused by Pb exposure.
Fig. 3: Proposed model of H3K27me3's roles in spatial memory deficits caused by Pb exposure.
Reference
- Gu X, Xu Y, Xue WZ, et al. Interplay of miR-137 and EZH2 contributes to the genome-wide redistribution of H3K27me3 underlying the Pb-induced memory impairment. Cell Death & Disease. 2019 Sep 11;10(9):671.


 Sample Submission Guidelines
Sample Submission Guidelines